Aldo Espinoza-Gómez a, c, Diana D. Moreno-Santillán d, Rafael Juárez-Maldonado a, Edgar G. Gutiérrez a, Ma. Isabel Salazar b, Luis A. Alonso-Palomares b, Juan Carlos López-Vidal c, Cynthia Elizalde-Arellano c, Jorge Ortega a, *
a Instituto Politécnico Nacional, Escuela Nacional de Ciencias Biológicas, Departamento de Zoología, Laboratorio de Bioconservación y Manejo, Posgrado en Ciencias Químicobiológicas, Prolongación de Carpio y Plan de Ayala s/n, Col. Santo Tomás, Miguel Hidalgo, 11340 Ciudad de México, Mexico
b Instituto Politécnico Nacional, Escuela Nacional de Ciencias Biológicas, Departamento de Microbiología, Laboratorio de Virología e Inmunovirología, Posgrado Ciencias Químicobiológicas, Prolongación de Carpio y Plan de Ayala s/n, Col. Santo Tomás, Miguel Hidalgo, 11340 Ciudad de México, Mexico
c Instituto Politécnico Nacional, Escuela Nacional de Ciencias Biológicas, Departamento de Zoología, Laboratorio de Cordados Terrestres, Prolongación de Carpio y Plan de Ayala s/n, Col. Santo Tomás, Miguel Hidalgo, 11340 Ciudad de México, Mexico
d Texas Tech University, Department of Biological Sciences, 2500 Broadway, Lubbock, Texas, 79409, USA
*Corresponding author: artibeus2@aol.com (J. Ortega-Reyes)
Received: 21 April 2021; accepted: 4 January 2022
Abstract
Bats are important reservoirs of viral entities that cause diverse economical and health problems in several sectors. The purpose of this study was to determine the prevalence of the rabies virus in vampire bats. We also tested for 3 other zoonotic viruses in the collected tissue. We captured 45 vampire bats (Desmodus rotundus), which were measured and sexed under standard parameters. We followed an RT-PCR reaction to amplify the viral products of four virus types: chikungunya (CHIKV), dengue (DENV), rabies (RABV) and Zika virus (ZIKV) from cerebral tissue. We obtained an amplicon of 100 bp for ZIKV in 2 samples, representing a prevalence of 4.4%. An amplicon of 581 bp for RABV was observed in 9 individuals, which is equivalent to 20% of our sample. We obtained 4 different haplotypes for RABV and a single haplotype for ZIKV. We did not record the presence of DENV and CHIKV. We corroborated the presence of the family Rhabdoviridae in the vampire bat and recorded the presence of ZIKV for the first time in this bat species.
Keywords: Chikungunya; Dengue; Flaviviridae; Rabies; Rhabdoviridae; Vampire bat; Zika
© 2022 Universidad Nacional Autónoma de México, Instituto de Biología. This is an open access article under the CC BY-NC-ND license
Identificación de secuencias de ARN viral en murciélagos vampiros (Desmodus rotundus) de México central
Resumen
Los murciélagos son reservorios importantes de virus que pueden causar diversos problemas económicos y de salud en diferentes sectores. El propósito de este estudio fue determinar la prevalencia del virus de la rabia en murciélagos vampiro. También se evaluó la prevalencia de 3 virus zoonóticos más en el tejido recolectado. Se capturó un total de 45 individuos, los cuales fueron medidos y sexados bajo parámetros estándar. Se efectuó la reacción de RT-PCR para amplificar los productos virales de 4 tipos de virus: chikungunya (CHIKV), dengue (DENV), rabia (RABV) y Zika (ZIKV) de tejido cerebral. Se obtuvo un amplicón de 100 pb para ZIKV en 2 muestras, lo que representa una prevalencia del 4.4%. Para el caso de RABV, se obtuvo un amplicón de 581 pb en 9 individuos, lo que equivale al 20% de la muestra. Se obtuvieron 4 haplotipos diferentes para RABV y un solo haplotipo para ZIKV. También se evaluó la presencia de virus del dengue (DENV) y virus de chikungunya (CHIKV), pero no se encontraron muestras positivas. Se corroboró la presencia de la familia Rhabdoviridae en el murciélago vampiro y se registró por primera vez la presencia de ZIKV en Desmodus rotundus.
Palabras clave: Chikungunya; Dengue; Flaviviridae; Rabia; Rhabdoviridae; Murciélago vampiro; Zika
© 2022 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
Introduction
Understanding why bats are major reservoirs of zoonotic viruses is an enduring objective in the field of virology. Co-evolutionary theory covers the close relationship between reservoirs/vectors and parasites; however, fine-scale details regarding the processes of pathogen acquisition, prevalence, host survival, and mechanisms of immunity are usually overlooked in many of the reports that describe the presence of viruses (Schountz, 2014). Detection of viruses in bats dates back to the middle of the last century when captured individuals were tested to establish prevalence. Many of the initial publications on the subject were limited to describing the recognition of a virus in a given species of bat (Calisher, 2015). Most studies showed incidental results regarding the detection of viruses without there being a particular objective for this type of study in bats. It was not until the 1980s that the field of virology began to gain relevance and to use more modern methods for virus detection (Calisher, 2015; Woo et al., 2006; Zhang et al., 1989). Recently, the detection of viruses by molecular methods has spread widely within the field. Current studies show viral sequences detected in bats, but it should be noted that these studies are reporting the nucleic acids of viral sequences and not the isolated virus per se. These viral sequences are only partial transcriptions of the viral genome that can be detected as endovirus in the host DNA, fragments, or complete viral genomes. Detection of these genomes by molecular techniques allows recognition of new pathogens in a host (Quan et al., 2013).
The wide diversity of viruses harbored by bats has been revealed by their extensive surveillance in the wild (Quan et al., 2013). These studies have determined a recent list of virus-bat relationships with a total number of 12 distinct viral families associated with the bats (Calisher, 2015; Calisher et al., 2006). The hematophagous vampire bat (Desmodus rotundus) is of great importance within the Chiroptera order since it is considered a species of relevance to public and veterinary health, particularly because of its propensity to spread rabies in livestock. Rabies transmitted by D. rotundus represents a risk to the health of cattle and its importance has been recognized in Latin America, where vampire bats are considered the main natural source of infection and reservoir of the etiological agent (Nadin-Davis & Loza, 2006; Velasco et al., 2006). The rabies virus (RABV) belongs to Lyssavirus (family Rhabdoviridae) and is transmitted via the saliva of infected animals through bites. Given their hematophagous habits, vampire bats usually cause serious epidemiological problems in livestock, in addition to considerable economic losses (Anderson et al., 2014). However, vampire bats can be asymptomatic reservoirs of viruses or can be productively infected with mortal consequences. In addition to rabies, vampire bats can be reservoirs of other viruses of human public health importance, such as some arbovirus belonging to the families Flaviviridae and Togaviridae (Kading & Schountz, 2016) that were included in this work (DENV, ZIKV, and CHIKV). Some studies support the evidence of infections in these bats; however, their relevance in the infection cycle is still unknown, which makes them an ideal ecological study model within the field of virology to elucidate their role as a potential reservoir for the dispersal of emerging viruses. Lack of studies on bat population in arbovirus such as ZIKV and CHIKV in Mexico supports the idea of carrying out new searches.
Viruses of the Flaviviridae and Togaviridae families are arthropod-borne since they require a blood-sucking arthropod to complete their cycle. It has been postulated that these viruses evolved in Africa, but they are now widely distributed across the tropical areas of the globe (Althouse et al., 2018). Outbreaks of dengue in the Americas have been reported since the 1800s, but more recent evaluations show that the disease is expanding its geographic area and infecting more people, mainly related to the increase in anthropogenic activities that cause alterations to the environment (Hernández-Aguilar et al., 2021; Petersen & Marfin, 2005). Zika disease has presented very rapid progress over recent decades, primarily in urban areas of Central America. Factors that have the greatest influence on the distribution of the vector within the aforementioned areas are correlated with optimum temperature points accompanied by ideal habitat conditions, which promote the population growth of the mosquito with an increase in the possible risk of disease transmission (Cunze et al., 2019). Chikungunya was reported for the first time in 2013 in the Caribbean French Island of Saint Martin, and has since spread to 45 countries of North, Central, and South America, with consecutive outbreaks occurring particularly in the rainy season. These countries have dealt with this new regional emergency in different ways, but the most effective measures for its management are associated with vector density and control (Yactayo et al., 2016). We selected these specific viruses because we wanted to understand the possible participation of these bats during the infection cycle, as a potential reservoir for these diseases of importance to humans, since there has been no evidence to date to positively support such a role.
To improve our knowledge regarding viruses of human health concern present in vampire bats, we assessed the prevalence of the viruses chikungunya (CHIKV), dengue (DENV), rabies (RABV), and Zika (ZIKV) in the brain tissue of the vampire bat. To fulfill our objective, we utilized molecular tools that allow us to detect sequences in brain tissues, but which do not necessarily indicate an active infection with these viruses.
Materials and methods
Four field trips were conducted from mid-2016 to mid-2017 in the Mexican states of Guanajuato, Puebla, and Yucatán, where vampire bats were reported (Elizalde-Arellano et al., 2010). We captured bats using mist nets of 6 m in length, set for 4 consecutive nights at the entrance of previously identified roosting sites for vampire bats (i.e., Socavón Cave in Guanajuato and Ejecayán Cave in Puebla). Hand nets were also used to collect individuals roosting during the day within the caves. Captured individuals were taxonomically identified using professional field guides (Medellín et al., 2008). These individuals were classified as adults when the wing epiphyses were completely ossified, and as juveniles when the joints were cartilaginous (Kunz, 1990), with standard morphological measurements taken to corroborate this classification. A spring scale (accurate to 0.1 g) was used to measure body mass, and an electronic caliper (accurate to 0.1 mm) was used to measure the total length and forearm length. We followed the bioethical norms of the American Society of Mammalogists, and the sampling procedures followed the ethical recommendations included in the Animal Care and Use Committee of the American Society of Mammalogists (Sikes et al., 2016). Collection permits were issued by the Secretaría de Medio Ambiente y Recursos Naturales, Mexico (SGPA/DGVS/02291/16, SGPA/DGVS/12772/17).
We performed cervical dislocation on each of the captured specimens and the brain was extracted from the skull and stored frozen in Eppendorf tubes with RNA later buffer until subsequent transportation to the laboratory. Each captured bat was handled individually, and precautionary measures were taken to decontaminate the extraction material by cleaning thoroughly with alcohol and soap each time the processing of the animal was finished. RNA from bat brains was extracted using the Trizol protocol (Chomczynsli & Sacchi, 1987) with 0.2 g of brain tissue. The RNA was re-suspended in H2O DEPC free of ribonuclease and stored at -70 °C. The RNA integrity was determined by 1% agarose gel electrophoresis stained with SYBR green® solution and with TAE 1X. Concentration of total RNA was quantified using a NanoDrop 2000 (Thermo Scientific TM Cat. No. ND-2000). For each RT-PCR reaction, we used a final concentration of 500 ng/μl of RNA.
Reverse Transcription-PCR (RT-PCR) endpoint technique was used with specific primers for the 4 screened viruses (DENV, CHIKV, RABV, and ZIKV) for amplification of the viral sequences, but a qPCR (Quantitative Polymerase Chain Reaction) to quantify viral expression was not used because we had no thermal cycler with fluorescence sensors that could give absolute quantification readings of the product. The PCR conditions and primers used are detailed in the supplementary material (Supplementary Material I: Table S1 to S5). The primers for each of the chosen viruses were selected according to their standard calibration use in medicine for diagnosis of the infection. These sequences are from highly conserved regions that allow an assertive diagnosis of the disease. The One-step RT-PCR kit (QIAGEN, Cat. No. /ID: 210212) was used to perform PCR amplification, according to the manufacturer’s specifications. All PCR products were visualized on 2% agarose gels stained with SYBR green®, using TBE 1X as a buffer for 40 minutes at 80 volts. We used a 50 bp ladder to assess the length of the amplified fragment. Positive controls for DENV, CHIKV, RABV, and ZIKV were obtained with the QIAamp Viral RNA kit (QIAGEN, Cat. No. /ID: 52904) from optimized source cultures isolated in the Virology and Immuno-Virology laboratory. RNA-free PCR was performed as a negative control (Chien et al., 2006).
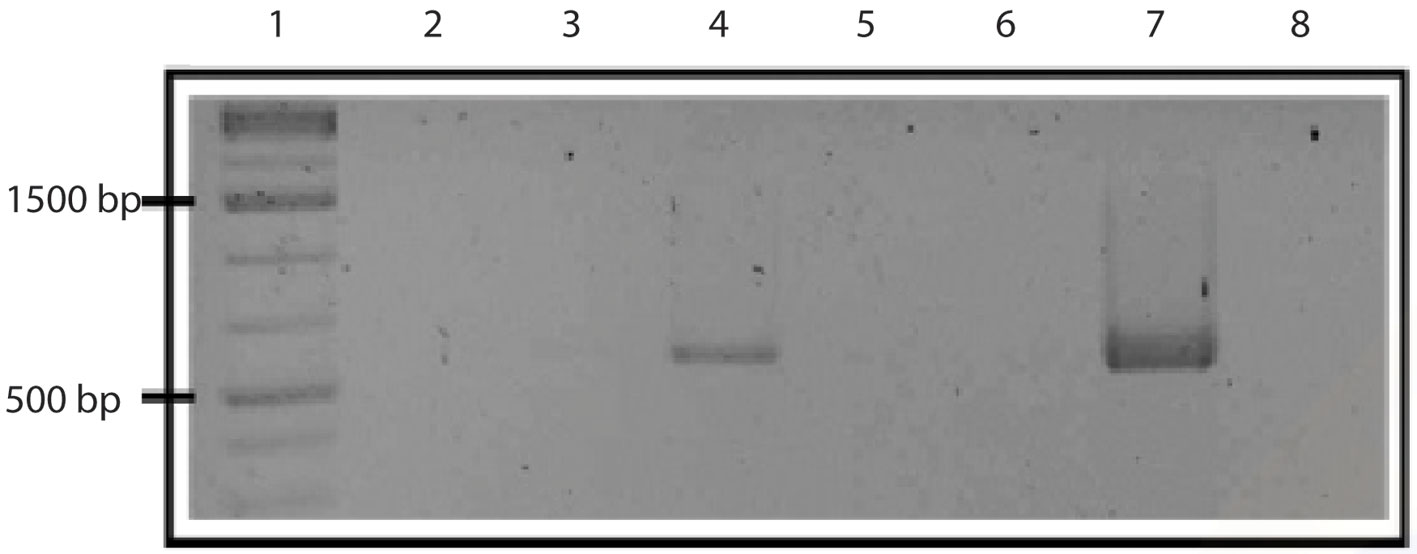
For detection of the rabies virus (RABV), a nested RT-PCR was used to increase the sensitivity and specificity of the amplification. This methodology was not applied to the other viruses because the standardized method does not detect nested PCR, according to the experience of the staff working in the Virology and Immuno-Virology laboratory of the university. A reaction was performed with the external primers to amplify a more extensive region of DNA, containing the target segment. The amplified product was then used as a template for a second PCR with internal primers to amplify a specific region containing the rabies virus sequence (an amplicon of 581 bp).
Positive samples were sent for sequencing to Macrogen, Inc. The obtained sequences were manually edited and aligned with BioEdit bioinformatics software (v 7.0.5). We queried sequences against the GenBank database employing BLAST (Altschul et al., 1990). A comparison between sequences was performed to detect differences in the nucleotide bases and determine the number of haplotypes present for each virus. We used BioEdit to translate nucleotide sequences into amino acid translations. We performed a posterior alignment among genotypes to determine synonymous and non-synonymous substitutions (Hall, 2001).
An evolutionary genotype network was obtained with the Free Phylogenetic Network Software (v.5) (Forster, 2015). We inferred ancestral types, potentially derived types, and evolutionary variants, as well as estimates of the number of mutations that had to have occurred to give rise to each genotype found. The network was constructed using only results with more than 2 genotypes; we completed the analysis with reference sequences from other strains obtained from the NCBI (National Center for Biotechnology Information – https://www.ncbi.nlm.nih.gov). We included GenBank sequences of the nucleoprotein N coding fragment for the RABV since this was our target fragment. However, only wildlife strains with this target exon that had been reported for Mexico were consulted in the database (Supplementary material II: Table S6). Those specific sequences were used to construct the genotype network (Fig. 3). The use of external genotypes presupposed and inferred the origin and diversification of our resulting strains circulating in Mexico.
Results
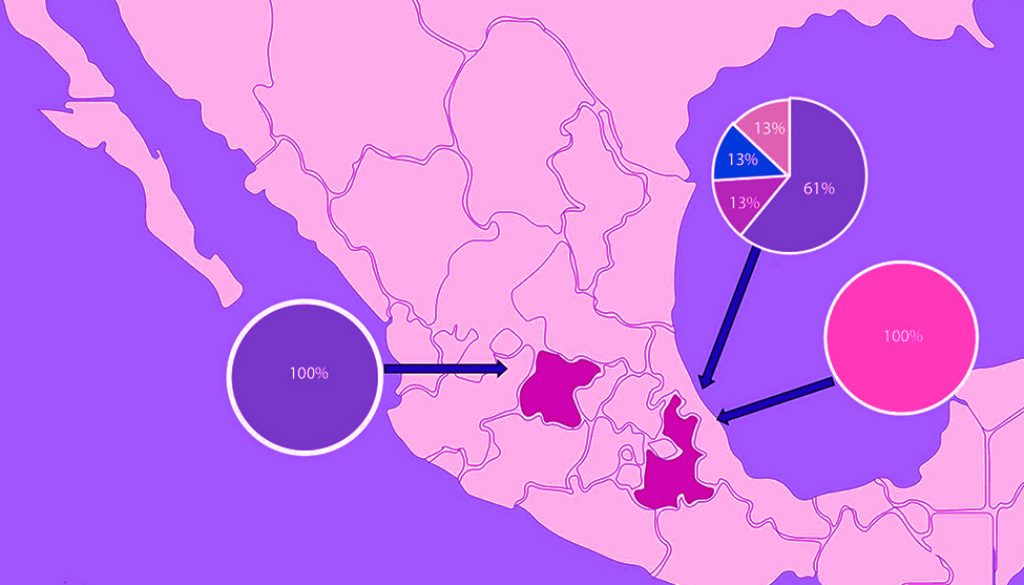
A total of 45 vampire bats were captured and sacrificed to extract the brain from the skull. We captured 8 males and 3 females in Guanajuato, 27 males and 5 females in Puebla, and 1 male and 1 female in Yucatán. All individuals were captured during the rainy season in Central Mexico and all of the captured individuals were adults and corroborated with the size of morphological measures corresponding to age and reported in the literature, whereas no juveniles were present in the colonies at that time of the year (Greenhall et al., 1983). A final amplicon of 581 bp was obtained in a positive RT-PCR test for RABV (Fig. 1). Positive RABV was found in 9 individuals (9/45 20% 95% CI 9-29): 7 adult males and 2 adult females. Eight positive detections belonged to vampire bats caught at the Ejecayán cave in Puebla, and 1 positive result came from the Socavón Cave in Guanajuato. We distinguished 4 different genotypes of rabies in the positive results (strains median vectors (mv) A to D). Guanajuato presented only the occurrence of genotype A, while Puebla exhibited all 4 described genotypes (Fig. 2). Synonymous and non-synonymous analyses are presented for each pairwise comparison among RABV genotypes (Table 1). Two non-synonymous mutational changes were found at positions 33 and 52, and 3 synonymous mutations at positions 7 and 39.
Table 1
Nucleotide position changes in the translation of amino acids for RABV. The synonymous changes are in bold, while the non-synonymous have asterisc.
| Position | ||||||||
| Genotype | 7 | 33 | 39 | 52 | ||||
| Nt | AA | Nt | AA | Nt | AA | Nt | AA | |
| A | A | Gln | A | Lys | T | Leu | T | Cys |
| B | G | Gln | A | Lys | T | Leu | G | Trp* |
| C | G | Gln | A | Lys | A | Leu | T | Cys |
| D | G | Gln | G | Arg* | A | Leu | T | Cys |
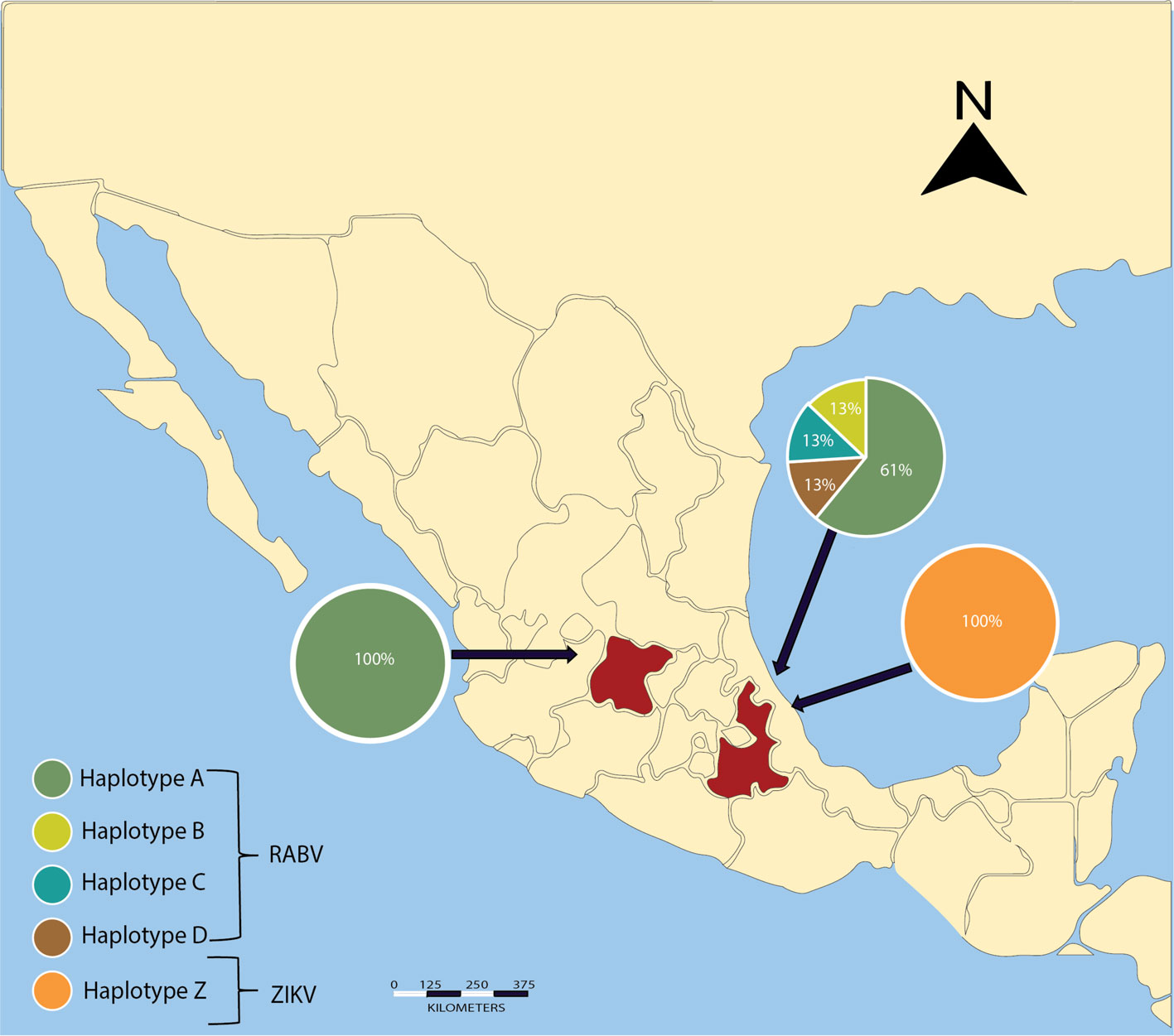
We were able to infer the genotype network only with RABV sequences. The grouping of the strains is represented by different colors (Fig. 3). All variants of RABV included in the genotype network are derived from the same theoretical ancestor (Median Vector 9 = mv9). We determined that genotypes B, C, and D came from the same hypothetical ancestor (mv1), which originated from genotype A (HTA). The size of genotype A is represented with a larger circle because of its abundance in our findings. Each putative ancestor represents a strain for which we have no sequence but is the only possible strain to give rise to each variant (Fig. 3).

CHIKV and DENV tested negative for all individuals at all localities. ZIKV tested positive for 2 individuals of Puebla (2/45 4.4%). A final amplicon of 100 bp is reported for the first time in vampire bats. No nucleotide differences were found among sequences of both individuals, which were considered a single genotype.
Discussion
Similar numbers of mammals and birds harbor zoonotic viruses (Mollentze & Stricker, 2020). Inconsistent results regarding virus isolation in bats have resulted in an overstated list of pathogens present in these animals. However, as mentioned in several studies, it is necessary to clarify whether these results refer to viral isolation (the gold standard for identification), or a simple genomic sequence detected by molecular analysis (Calisher, 2015). Establishing whether only viral genomic sequences are obtained, rather than identifying the complete virus, is important to reduce the degree of error in identification, since the gold standard protocol has a sensitivity of 100% in terms of identifying the disease, while partial genomic sequences may be subject to a first mismatch. These discrepancies between methodologies can foster the negative reputation for the bats as virus reservoirs and can lead to the implementation of governmental actions that negatively impact bat populations. Detection of a viral sequence in a bat does not necessarily imply a viral infection cycle; some animals act as dead-end hosts, and individuals may act as reservoirs or present an asymptomatic form of the disease. There is a grey area of information on the mechanisms of infection, forms of dispersion, pathogenicity, and occurrence of diseases in wild mammals that obscures the co-evolutive importance of the relationship (Streicker et al., 2016). Rodents and bats are the taxonomic groups that harbor the most zoonotic viruses, and this seems a consequence of their diversity and species richness (Johnson et al., 2020). Killing bats or destroying their natural habitats may be more likely to increase the occurrence of these viruses in humans, and spread from bat reservoirs can occur even when bats are asymptomatic (Johnson et al., 2020). Result is a direct consequence of human actions towards nature (e.g., habitat destruction), although only a few cases of recorded spillover have caused problems for humans.
Blood collection was not considered during our sampling because initially we only intended to estimate the seroprevalence of RABV in brain tissue; however, the seroprevalence of flavivirus in serum had been shown in Neotropical bats (Machain-Williams et al., 2013). We took the opportunity to test for other viruses in brain tissue, even though it is not the most assertive material, based on scientific evidence about how specific viral infections can cause neurological complications, severe brain damage, and the development of acute syndromes with ZIKV (Zunk, 2017), CHIKV (Agarwal et al., 2017; Kumar et al., 2019; van den Pol et al., 2017), and DENV (Calderón-Peláez et al., 2019; Velandia-Romero et al., 2016).
Vampire bats have been recognized as a major reservoir host for rabies and are a constant threat for rabies transmission to livestock, which causes important losses and economic impact. Vampire bat abundance and population dynamics may have consequences in infectious disease transmission; due to this population trend, monitoring their populations should be a novel approach to mitigate rabies risk transmitted by bats (Anderson et al., 2014; Gonçalves et al., 2021). We identified the prevalence of rabies virus strains in vampire bats from 2 locations and included 3 members of other major viruses in our analysis because the role of this bat as a potential virus reservoir could be of great interest in terms of human health concerns related to emerging diseases; however, more studies need to be evaluated (Abundes-Gallegos et al., 2018; Fagre & Kading, 2019). The importance of this study lies in the use of molecular methods for the detection of viruses in wildlife, in addition to a novel approach to the analysis of the data.
Rabies is one of the most life-threatening diseases that occur at the zoonotic level in both wildlife and urban populations. Numerous incidents of rabies in human populations, transmitted by a vampire bat bite, have been documented (Cifuentes-Jiménez et al., 2017; Da Rosa et al., 2006; Dato et al., 2016). Moreover, the cost associated with the transmission of rabies to livestock is high in terms of economic losses and necessitates continuous vaccination campaigns, as well as the eradication of neighboring vampire bat colonies (Anderson et al., 2014). The pathogenesis of the disease has been addressed in various ways for bats (Aguilar-Setien et al., 2002; Ellison et al., 2013) but, in practical terms, many governmental decisions are based on eradicating entire populations of vampire bats with no prior research conducted to establish the prevalence of the disease in these animals, and the mitigating effect of these bat eradication programs in terms of reducing RABV incidence remains to be adequately studied. Culling adult vampire bats is not an effective disease control measure, considering that the seroprevalence in a colony of approximately 500 individuals is 20% and is particularly high in juvenile individuals (Streicker et al., 2012). In general, seroprevalence is often high in bat colonies, but viral prevalence is low when measured in individuals. Our finding of a high incidence of RABV is exceptional compared with the usual findings in other studies (Streicker et al., 2012). Then, we pointed to maximum care in the treatment of the samples to avoid cross-contamination.
The prevalence of rabies virus in vampire bats has been documented, but poorly analyzed. Many papers seek to demonstrate the seroprevalence of the virus in the species and its spread throughout its geographic distribution in Mexico (Johnson et al., 2014). Livestock such as cows, pigs, horses, among others, represent an important food source for vampire bats; it has even been documented that their food preference is skewed towards cattle over other potential wildlife resources (Johnson et al., 2014; Stoner-Duncan et al., 2014). The demand for non-native food resources by vampire bats is closely related to changes in land-use, which beneficially alter its foraging mechanism and promote greater spread of rabies among livestock (Streicker & Allgeiger, 2016). This epidemiological characteristic of transmitting rabies to livestock makes it possible to consider vampire bats as an important ecological vector, the dynamics of which must be considered with great care to prevent the spread of the disease while adequately managing their populations.
The differences in genotypic diversity found between the 2 provinces in this study may be the result of the wide distribution of the vampire bats in Mexico, combined with the geographical distance that separates the 2 sites. The results of the genotype network indicate that genotype A is the most abundant, as well as being the common ancestor for the rest of the derived genotypes, which may be indicative of a successful genotype due to its active persistence. The discovery of a series of variable genotypes in a single locality suggests the occurrence of more opportunities for the adaptability of genotypes due to host and transmission routes (Zhang et al., 2003). The comparisons between the sequences obtained from GenBank and our genotypes showed dissimilarities due to a few mutational changes but indicate the existing strains of circulating rabies in Mexico that are affecting different groups of mammals. The distinct groups of genotypes shown in figure 3 show a certain degree of similarity while presenting a few mutational steps between them, reflecting recent evolutionary derivations. The genotypes found for vampire bats in Mexico and reported in this study increase the number of circulating strains by 36% and complement those reported in GenBank (only for the nucleoprotein N sequence). Non-synonymous mutations can cause structural and functional changes in the protein if the encoding of the amino acid acts to change its functionality. We found 2 non-synonymous mutations in a RABV genotype but are unaware of the impact these may have on the virus, considering it is merely a fragment of the amplified protein.
In our results, the diversity of viral sequences was low for ZIKV and null for CHIKV and DENV, considering the neurotropism of these viruses, we evaluated brain tissue; however, we did not evaluate other tissues that may be a potential target of arboviral infections. ZIKV was isolated for the first time in West Africa in the mid-1990s and found more recently in Brazil in 2013 (Bueno et al., 2016). Few studies have focused on the role of animals as hosts for ZIKV, but several studies have reported ZIKV antibodies in different mammalian species (e.g., primates, ungulates, carnivores, rodents, and proboscideans; Bueno et al., 2016; Chan et al., 2016). We reported for the first time the occurrence of a DNA fragment of ZIKV in vampire bats; however, the presence of positive individuals does not imply a current zoonotic development and potential transmission, thus further studies must be conducted to establish the true potential in this regard. Positive sequences of ZIKV have been identified in non-human Neotropical primates, and laboratory experiments in marmosets showed sustained viremia in treated animals (Terzian et al., 2018). A recent expansion of the virus in Latin America and the infinite possibility of hosts corresponding to wildlife could lead to an increase in cases of this emerging disease (Moser et al., 2018; Shaily & Upadhya, 2019).
The coevolution between ZIKV and its main vector, Aedes aegypti, has been studied during the most recent epidemics of this worldwide disease, but scientists have no actual evidence to suggest that the mosquito feeds on vampire bats (Anderson et al., 2016; Weissenböck et al., 2010). The outbreaks and their rapid expansion in Latin America can be explained by a continuous infection of other mammalian hosts, in addition to active infection of the widely distributed vectors or reservoirs. It has been proposed that a non-structural protein can more easily facilitate the acquisition of ZIKV through the blood consumed by the mosquito. In habitats where human populations are scarce, mammals could become the main hosts and could maintain ZIKV from season to season or infect mosquito vectors. High mosquito abundance and the presence of reservoirs pose a risk of new outbreaks and reinfections, as well as a permanent prevalence of the disease in risk areas (Malmlov et al., 2019; Noor & Ahmed, 2018). Experimental infections in bats with ZIKV showed no viremia development in the manipulated animals and epidemiological studies are therefore necessary to establish the role of wildlife in the functionality of the disease (Fagre & Kading, 2019).
Experimental infections with CHIKV in wildlife in the USA produced no detectable viremia or overt clinical disease in 9 avian and 12 mammalian species, aside from the big brown bats (Eptesicus fuscus). These animals belong to a distinct geographical area where the virus was first detected, and the infection is considered a novel disease. A major conclusion was no significant threat of CHIKV to the wildlife and a null contribution of the biota to the natural transmission cycle of the virus (Bosco-Lauth et al., 2016). In geographical areas where CHIKV has evolved and caused numerous outbreaks, positive serological samples were detected in simian primates, but null results were found in other mammalian species (Althouse et al., 2018; Higgs & Ziegler, 2010; Vourc´h et al., 2014). CHIKV is primarily maintained in Africa and Asia via a zoonotic sylvatic cycle that relies on nonhuman primates as reservoirs (Tsetsarkin et al., 2011).
Serological studies have suggested a sylvatic cycle of DENV in wildlife. Low proportions of DENV circulating in wildlife have shown the low incidence of the disease in these organisms, including circulating strains that differ between those found in different groups of mammals and those present in urban populations (Calderón et al., 2019; Hernández-Aguilar et al., 2021; Lavergne et al., 2009). The strains of DENV that circulate in human populations are highly infectious and present a poor immunological response, while those detected in wild animals show low seroprevalence (Pandit et al., 2018; Raut et al., 2019). In our study none of the brain tissues sampled was positive for DENV, with no clear relationship with the disease and potential interspecific transmission. Experimental infections with bats using DENV serotypes showed low levels of viremia, or development of symptomatology of the disease, with a lack of detection of viral RNA in the organs via RT-PCR (Fagre & Kading, 2019). Multiple experimental infections showed the same results, confirming that replication of DENV in the host is null or scarce, with a consequently low probability of constituting a reservoir of the disease (Fabre & Kading, 2019).
However, viral arrangements of DENV have been detected previously in vampire bat tissues of central Mexico, and ectoparasites inherent to these bats have presented Flaviviridae sequences (Abundes-Gallegos et al., 2018). Seroprevalence analysis of DENV and PCR showed no significant findings of disease or infection in Neotropical bats associated with households, suggesting an incidental form of the presence of DENV in these mammals (Vicente-Santos et al., 2017). However, laboratory-induced infections with bats showed tissue damage and the development of symptoms associated with dengue (Perea-Martínez et al., 2013).
There is much uncertainty regarding how climate change can affect the distribution, abundance, and prevalence of many diseases, especially those of tropical origin, which could affect human populations in the area, whereas this topic must be studied deeply to understand the real impact of global warming (Colón-González et al., 2013). In our study, we analyzed the incidence of 4 emerging and human-related diseases in vampire bats of 3 localities of Mexico, a species that benefits from anthropogenic disturbances and has the potential to increase its range of distribution by adjusting to environmental changes (Piaggio et al., 2017; Streicker et al., 2016). Our results show low numbers of positivity at 25% for rabies, which coincides with other prevalence studies (Escobar et al., 2015). The incidence of ZIKV in vampire brain tissue is also reported for the first time, which is a novel result in this study. Moreover, we detected no incidence of 2 other tropical diseases. It is clear that detection of diseases in wildlife is challenging, but this type of research can further our understanding of wildlife as a reservoir of viruses and its possible future implications for both human health and the maintenance of bat populations.
Acknowledgments
Financial and material support was provided by Conacyt Ciencia Básica 156725 and by SIP-IPN 20190022 granted by the Instituto Politécnico Nacional to J. Ortega and by SIP-IPN 20160096 granted by the Instituto Politécnico Nacional to C. Elizalde-Arellano. We also acknowledge JE Muñoz-Medina from the Central Laboratory of Epidemiology at the Instituto Mexicano del Seguro Social for providing Arbovirus primers, and A. Aguilar-Setien from the Unidad de Investigación Médica en Inmunología, Centro Médico Nacional Siglo XXI, for providing Rabies virus primers.
References
Abundes-Gallegos, J., Salas-Rojas, M., Gálvez-Romero, G., Perea-Martínez, L., Obregón-Morales, C. Y., Morales-Malacara, J. B. et al. (2018). Detection of dengue virus in bat flies (Diptera: Streblidae) of common vampire bats, Desmodus rotundus, in Progreso, Hidalgo, Mexico. Vector-Borne and Zoonotic Diseases, 18, 70–73.
Agarwal, A., Vibha, D., Srivastava, A. K., Shukla, G., & Prasad, K. (2017). Guillain-Barre syndrome complicating chikungunya virus infection. Journal of Neurovirology, 23, 504–507. https://doi.org/10.1007/s13365-017-0516-1
Aguilar-Setien, A., León Campos, Y., Tesoro-Cruz, E., Kretschmer, R., Brochier, B., & Pastoret, P. R. (2002). Vaccination of vampire bats using recombinant vaccinia-Rabies virus. Journal of Wildlife Diseases, 38, 539–544. https://doi.org/10.7589/0090-3558-38.3.539
Althouse, B. M., Guerbois, M., Cummings, D. A. T., Diop, O. M., Faye, O., Faye, A. et al. (2018). Role of monkeys in the sylvatic cycle of chikungunya virus in Senegal. Nature Communications, 9, 1046.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Anderson, A., Shwiff, S. S., & Shwiff, S. A. (2014). Economic impact of the potential spread of vampire bats into South Texas. In R. M. Timm, & J. M. O’Brien (Eds.), Proceedings 26th Vertebrate Pest Conference (pp. 305–309). University of California, USA.
Anderson, K. B., Thomas, S. J., & Endy, T. P. (2016). The emergence of Zika virus. A narrative review. Annals of Internal Medicine, 165, 175–183. https://doi.org/10.7326/M16-0617
Bobrowiec, P. E. D., Lemes, M. R., & Brigel, R. (2015). Prey preference of the common vampire bat (Desmodus rotundus, Chiroptera) using molecular analysis. Journal of Mammalogy, 96, 54–63.
Bosco-Lauth, A., Nemeth, N. M., Kohler, D. J., & Bowen, R. A. (2016). Viremia in North American mammals and birds after experimental infection with Chikungunya viruses. American Journal of Tropical Medicine Hygiene, 94, 504–506. https://doi.org/10.4269/ajtmh.15-0696
Bueno, M. G., Martínez, N., Abdalla, L., Duarte Dos Santos, C. N., & Chame, M. (2016). Animals in the Zika Virus life cycle: What to expect from megadiverse Latin American countries. Plos Neglected Tropical Diseases, 10, e0005073. https://doi.org/10.1371/journal.pntd.0005073
Calderón, A., Guzmán, C., Mattar, S., Rodríguez, V., Martínez, C., Violet, L. et al. (2019). Dengue virus in bats from Córdoba and Sucre, Colombia. Vector Borne and Zoonotic Diseases, 19, 747–751. https://doi.org/10.1089/vbz.2018.2324
Calderón-Peláez, M. A., Velandia-Romero, M. L., Bastidas-Legarda, L. Y., Beltrán, E. O., Camacho-Ortega, S. J., & Castellanos, J. E. (2019). Dengue virus infection of blood-brain barrier cells: consequences of severe disease. Frontiers in Microbiology, 10, 1435. https://doi.org/10.3389/fmicb.2019.01435
Calisher, C. H. (2015). Viruses in bats: a historic review. In L. F. Wang, & C. Cowled (Eds.), Bats and viruses: a new frontier of emerging infectious diseases (pp. 23–45). New Jersey: John Wiley & Son.
Calisher, C. H., Childs, J. E., Field, H. E., Holmes, K. V., & Schountz, T. (2006). Bats: important reservoir hosts of emerging viruses. Clinical Microbiology Reviews, 19, 531–545. https://doi.org/10.1128/CMR.00017-06
Chan, J. F. W., Choi, G. K. Y., Yip, C. C. Y., Cheng, V. C. C., & Yuen, K.-Y. (2016). Zika fever and congenital Zika syndrome: An unexpected emerging arboviral disease. Journal of Infection, 72, 507–524. https://doi.org/10.1016/j.jinf.2016.02.011
Chien, L. J., Liao, T. L., Shu, P. Y., Huang, J. H., Gubler, D. J., & Chang, G. J. (2006). Development of real-time reverse transcriptase PCR assays to detect and serotype dengue viruses. Journal of Clinical Microbiology, 44, 295–304. https://doi.org/10.1128/JCM.44.4.1295-1304.2006
Chomczynsli, P., & Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry, 161, 156–159.
Cifuentes-Jiménez, J. F., Pérez-López, R. D., & Verjan-García, N. (2017). Bat reservoirs for Rabies virus and epidemiology of rabies in Colombia: a review. Revista CES Medicina Veterinaria Zootecnista, 12, 134–150. https://doi.org/10.21615/cesmvz.12.2.5
Colón-González, F. J., Fezzi, C., Lake, I., & Hunter, P. R. (2013). The effects of weather and climate change on Dengue. Plos Neglected Tropical Diseases, 7, e2503. https://doi.org/10.1371/journal.pntd.0002503
Cunze, S., Kochmann, J., Koch, L. K., Genthner, E., & Klimpel, S. (2019). Vector distribution and transmission risk of the Zika virus in South and Central America. PeerJ, 7, e7920. https://doi.org/10.7717/peerj.7920
Da Rosa, E. S. T., Kotatit, I., Barbosa, T. F. S., Carrieri, M. L., Brandão, P. E., Pinheiro, A. S. et al. (2006). Bat-transmitted human Rabies outbreaks, Brazilian Amazon. Emerging Infectious Diseases, 12, 1197–1202. https://doi.org/0.3201/1208.050929
Dato, V. M., Campagnolo, E. R., Long, J., & Rupprecht, C. R. (2016). A systematic review of human bat Rabies virus variant cases: Evaluating unprotected physical contact with claws and teeth in support of accurate risk assessments. Plos One, 11, e0159443. https://doi.org/10.1371/journal.pone.0159443
Elizalde-Arellano, C., López-Vidal, J. C., Uhart, E. Q., Campos-Rodríguez, J. I., & Hernández-Arciga, R. (2010). Nuevos registros y extensiones de distribución de mamíferos para Guanajuato, México. Acta Zoológica Mexicana, 26, 73–98. https://doi.org/10.21829/azm.2010.261681
Ellison, J. A., Johnson, S. R., Kuzmina, N., Gilbert, A., Carson, W. C., Vercauteren, K. C. et al. (2013). Multidisciplinary approach to epizootiology and pathogenesis of bat Rabies viruses in the United States. Zoonoses and Public Health, 60, 46–57. https://doi.org/10.1111/zph.12019
Escobar, L. E., Townsend-Peterson, A., Favi, M., Yung, V., & Medina-Vogel, G. (2015). Bat-borne Rabies in Latin America. Revista del Instituto de Medicina Tropical de Sao Paulo, 57, 63–72. https://doi.org/10.1590/S0036-46652015000100009
Fagre, A. C., & Kading, R. C. (2019). Can bats serve as reservoirs for Arboviruses? Viruses, 11, 215. https://doi.org/10.3390/v11030215
Forster, M. (2015). Network 5.0.0.0 User Guide. In Fluxus Technology 2020. Network 10. Consulted January 08, 2019 of: www.fluxus-engineering.com/Network5000 _user_guide.pdf
Gonçalves, F., Galetti, M., & Streicker, D. (2021). Management of vampire bats and rabies: a precaution for rewilding projects in the Neotropics. Perspectives in Ecology and Conservation, 19, 37–42. https://doi.org/10.1016/j.pecon.2020.12.005
Greenhall, A. M., Joermann, G., & Schmidt, U. (1983). Desmodus rotundus. Mammalian Species, 202, 1–6. https://doi.org/10.2307/3503895
Hall, T. (2001). BioEdit v5.0.6 Program distributed by the author. Department of Microbiology. North Carolina State University. North Carolina, USA.
Hernández-Aguilar, I., Lorenzo, C., Santos-Moreno, A., Navarrete Gutiérrez, D., & Naranjo, E. J. (2021). Current knowledge and ecological and human impact variables involved in the distribution of the dengue virus by bats in the Americas. Vector-Borne and Zoonotic Diseases, 21, 217–231. https://doi.org/10.1089/vbz.2020.2696
Higgs, S., & Ziegler, S. A. (2010). A nonhuman primate model of chikungunya disease. The Journal of Clinical Investigation, 120, 657–660. https://doi.org/10.1172/JCI42392
Johnson, C. K., Hitchens, P. L., Pandit, P. S., Rushmore, J., Evans, T. S., Young, C. C. W., & Doyle, M. M. (2020). Global shifts in mammalian population trends reveal key of virus spill over risk. Proceeding of the Royal Society B, 287, 20192736. https://doi.org/10.1098/rspb.2019.2736
Johnson, N., Aréchiga-Ceballos, N., & Aguilar-Setien, A. (2014). Vampire bat rabies: Ecology, epidemiology and control. Viruses, 6, 1911–1928. https://doi.org/10.3390/v6051911
Kading, R. C., & Schountz, T. (2016). Flavivirus infections of bats: potential role in Zika virus ecology. The American Journal of Tropical Medicine and Hygiene, 95, 993–996. https://doi.org/10.4269/ajtmh.16-0625
Kumar, R., Rajvanshi, P., Khosla, H., & Aroa, S. (2019). Neuro-Chikungunya: acute transverse myelopathy associated with Chikungunya virus infection. The Journal of the Association of Physicians of India, 67, 84–85.
Kunz, T. H. (1990). Ecological and behavioural methods for the study of bats. Washington D.C.: Smithsonian Institution.
Kuzmin, I. V., & Rupprecht, C. E. (2015). Bat Lyssaviruses. In L. F. Wang, & C. Cowled (Eds.), Bats and viruses: a new frontier of emerging infectious diseases (pp. 47–97). New Jersey: John Wiley & Son.
Lavergne, A., Lacoste, V., Germain, A., Matheus, S., Dussart, P., Deparis, X. et al. (2009). Dengue virus infection in neotropical forest mammals: Incidental hosts or potential reservoirs? Médecine Tropicale: Revue du Corps de Santé Colonial, 69, 345–350.
Machain-Williams, C., López-Uribe, M., Talavera-Aguilar, L., Carrillo-Navarrete, J., Vera-Escalante, L., Puerto-Manzano, F. et al. (2013). Serologic evidence of Flavivirus infection in bats in the Yucatán Peninsula of Mexico. Journal of Wildlife Disease, 49, 684–689. https://doi.org/10.7589/2012-12-318
Malmlov, A., Bantle, C., Aboellail, T., Wagner, K., Campbell, C. L., Eckley, M. et al. (2019). Experimental Zika virus infection of Jamaican fruit bats (Artibeus jamaicensis) and possible entry of virus into brain via activated microglial cells. Plos Neglected Tropical Diseases, 13, e0007071. https://doi.org/10.1371/journal.pntd.0007071
Medellín, R. A., Arita, H., & Sánchez, O. (2008). Identificación de los murciélagos de México, clave de Campo. Ciudad de México: Instituto de Ecología, Universidad Nacional Autónoma de México.
Mollentze, N., & Streicker, D. G. (2020). Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. PNAS, 117, 9423–9430. https://doi.org/10.1073/pnas.1919176117
Moser, L. A., Boylan, B. T., Moreira, F. R., Myers, L. J., Svenson, E. L., Fedorova, N. B. et al. (2018). Growth and adaptation of Zika virus in mammalian and mosquito cells. Plos Neglected Tropical Diseases, 12, e0006880. https://doi.org/10.1371/journal.pntd.0006880
Nadin-Davis, S. A., & Loza, E. (2006). The molecular epidemiology of rabies associated with chiropteran hosts in Mexico. Virus Research, 117, 215–226. https://doi.org/10.1016/j.virusres.2005.10.006
Noor, R., & Ahmed, T. (2018). Zika virus: Epidemiological study and its association with public health risk. Journal of Infection and Public Health, 11, 611–616. https://doi.org/10.1016/j.jiph.2018.04.007
Pandit, P. S., Doyle M. M., Smart, K. M., Young C. C. W., Drape G. W., & Johnson C. K. (2018). Predicting wildlife reservoirs and global vulnerability to zoonotic Flaviviruses. Nature Communications, 9, 5425. https://doi.org/10.1038/s41467-018-07896-2
Perea-Martínez, L., Moreno-Sandoval, H. N., Moreno-Altamirano, M. M., Salas-Rojas, M. M., García-Flores, M. M., Aréchiga-Ceballos, N. et al. (2013). Experimental infection of Artibeus intermedius bats with serotype-2 dengue virus. Comparative Immunology, Microbiology & Infectious Diseases, 36, 193–198. https://doi.org/10.1016/j.cimid.2012.12.002
Petersen, L. R., & Marfin, A. A. (2005). Shifting epidemiology of Flaviviridae. Journal of Travel Medicine, 12, S3–S11. https://doi.org/10.2310/7060.2005.12052
Piaggio, A. J., Russell, A. L., Osorio, I. A., Jiménez-Ramírez, A., Fischer, J. W., Neuwald, J. L. et al. (2017). Genetic demography at the leading edge of the distribution of a rabies virus vector. Ecology and Evolution, 7, 1–9. https://doi.org/10.1002/ece3.3087
Premawansa, S., Wijewickrama, A., Premawansa, G. et al. (2019). Dengue type 1 viruses circulating in humans are highly infectious and poorly neutralized by human antibodies. Proceedings of the National Academy of Sciences, 116, 227–232. https://doi.org/10.1073/pnas.1812055115
Quan, P. L., Firth, C., Conte, J. M., Williams, S. H., Zambrana-Torrelio, C. M., Anthony, S. J. et al. (2013). Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proceedings of the National Academy of Sciences, 110, 8194–8199. https://doi.org/10.1073/pnas.130303711
Raut, R., Corbett, K. S., Tennekoon, R. N., Romero-Nava, C., León-Paniagua, L., & Ortega, J. (2014). Microsatellite loci reveal heterozygosis and population structure in vampire bats (Desmodus rotundus) (Chiroptera: Phyllostomidae) of Mexico. International Journal of Tropical Biology and Conservation, 62, 659–669.
Schountz, T. (2014). Immunology of bats and their viruses: Challenges and opportunities. Viruses, 6, 4880–4901. https://doi.org/10.3390/v6124880
Shaily, S., & Upadhya, A. (2019). Zika virus: Molecular responses and tissue tropism in the mammalian host. Reviews in Medical Virology, 29, e2050. https://doi.org/10.1002/rmv.2050
Sikes, R. S., & the Animal Care and Use Committee of the American Society of Mammalogists. (2016). Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. Journal of Mammalogy, 97, 663–688. https://doi.org/10.1093/jmammal/gyw078
Stoner-Duncan, B, Streicker, D. G., & Tedeschi, C. M. (2014). Vampire bats and Rabies: Toward an ecological solution to a public health problem. Plos Neglected Tropical Diseases, 8, e2867. https://doi.org/10.1371/journal.pntd.0002867
Streicker, D. G., & Allgeiger, J. E. (2016). Foraging choices of vampire bats in diverse landscapes: potential implications for land-use change and disease transmission. Journal of Applied Ecology, 53, 1280–1288. https://doi.org/10.1111/1365-2664.12690
Streicker, D. G., Recuenco, S., Valderrama, W., Gómez-Benavides, J., Vargas, I., Pacheco, V. et al. (2012). Ecological and anthropogenic drivers of rabies exposure in vampire bats: Implications for transmission and control. Proceedings of the Royal Society B, 279, 3384–3392. https://doi.org/10.1098/rspb.2012.0538
Streicker, D. G., Winternitz, J. C., Satterfield, D. A., Condori-Condori, R. E., Broos, A., Tello, C. et al. (2016). Host-pathogen evolutionary signatures reveal dynamics and future invasions of vampire bat rabies. Proceedings of the National Academy of Sciences, 113, 10926–10931. https://doi.org/10.1073/pnas.160658711
Terzian, A. C. B., Zini, N., Saccheto, L., Rocha, R. F., Parra, M. C. P., Del Sarto, J. L. et al. (2018). Evidence of natural Zika virus infection in neotropical non-human primates in Brazil. Scientific Reports, 8, 16034. https://doi.org/10.1038/s41598-018-34423-6
Tsetsarkin, K. A., Chen R., Leal G., Forrester N., Higgs S., Huang J. et al. (2011). Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proceeding of the National Academy of Sciences, 108, 7872–7877. https://doi.org/10.1073/pnas.1018344108
van Den Pol, A. N., Mao, G., Chattopadhyay, A., Rose, J. K., & Davis, J. N. (2017). Chikungunya, Influenza, Nipah, and Semliki forest chimeric viruses with vesicular stomatitis virus: Actions in the brain. Journal of Virology, 91, e02154–16. https://doi.org/10.1128/JVI.02154-16
Velandia-Romero, M., Calderón-Peláez, M. A., & Castellanos, J. E. (2016). In Vitro infection with dengue virus changes in the structure and function of the mouse brain endothelium. Plos One, 11, e0157786. https://doi.org/10.1371/journal.pone.0157786
Velasco, V. A., Orciari, L. A., Juárez, V., Gómez, M., Padilla, I., & Flisser, A. (2006). Molecular diversity of rabies associated with bats in Mexico and other countries of the Americas. Journal of Clinical Microbiology, 44, 1697–1710. https://doi.org/10.1128/JCM.44.5.1697-1710.2006
Vicente-Santos, A., Moreira-Soto, A., Soto-Garita, C., Chaverri, L. G., Chaves, A., Drexler, J. F. et al. (2017). Neotropical bats that co-habit with human function as dead-end hosts for dengue virus. Plos Neglected Tropical Diseases, 11, e0005537. https://doi.org/10.1371/journal.pntd.0005537
Vourc´h, G., Halos, L., Desvars, A., Boué F., Pascal M., Lecollinet S. et al. (2014). Chikungunya antibodies detected in non-human primates and rats in three Indian Ocean islands after the 2006 ChikV outbreak. Veterinary Research, 45, 52.
Weissenböck, H., Hubálek, Z., Bakonyi, T., & Nowtny, N. (2010). Zoonotic mosquito-borne flaviviruses: Worldwide presence of agents with proven pathogenicity and potential candidates of future emerging diseases. Veterinary Microbiology, 140, 271–280. https://doi.org/10.1016/j.vetmic.2009.08.025
Woo, P. C., Lau, S. K., Li, K. S., Poon, R. W., Wong, B. H., Tsoi, H. W. et al. (2006). Molecular diversity of coronaviruses in bats. Virology, 351, 180–187. https://doi.org/10.1016/j.virol.2006.02.041
Yactayo, S., Staples, J. E., Millot, V., Cibrelus, L., & Ramon-Pardo, P. (2016). Epidemiology of Chikungunya in the Americas. The Journal of Infectious Diseases, 214, S441–S445. https://doi.org/10.1093/infdis/jiw390
Zhang, H. L., Shi, H. F., & Liu, L. H. (1989). Isolation of chikungunya virus from bat in Yunnan province and serological investigations. Chinese Journal of Virology, 5, 31–36. https://doi.org/10.1371/journal.pone.0061950
Zhang, J., Rowe, W. L., Clark, A. G., & Buetow, K. H. (2003). Genome wide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. American Journal of Human Genetics, 73, 1073–1081. https://doi.org/10.1086/379154
Zunk, J. R. (2017). Neurologic manifestations of Zika virus infection. Anales de la Facultad de Medicina, 8, 83–87.