Genetic analysis of wild drone congregations of the stingless bee Scaptotrigona mexicana (Hymenoptera: Apidae) reveals a high number of colonies in a natural protected area in Southern Mexico
Daniel Sánchez a, *, Rémy Vandame a, Frank B. Kraus b, c
a El Colegio de la Frontera Sur, Km 2.5 Carretera Antiguo Aeropuerto, 30700 Tapachula, Chiapas, Mexico
b Institut für Biologie,Martin-Luther-Universität Halle-Wittenberg, HoherWeg 4, D-06099 Halle (Saale), Germany
c Department of Laboratory Medicine, University Hospital Halle (Saale), Ernst-Grube Str. 40 Halle (Saale), Germany
* Corresponding author: dsanchez@ecosur.mx (D. Sánchez)
Abstract
In this study, drone congregations of the stingless bee Scaptotrigona mexicana Guérin were genetically analyzed at 5 microsatellite loci to estimate the number of colonies of this species in the La Sepultura Biosphere Reserve, in Chiapas, Mexico. A total of 216 drones from 3 congregations were analyzed. We found that the 3 congregations had enough genetic differences to be considered as coming from different populations. The average number (± SD) of colonies per congregation was 19.8 ± 0.63, 17.6 ± 0.70 and 12. A similar result was reported for areas where stingless beekeeping occurs. We discuss the implications of our findings in a conservation framework in which stingless beekeeping might play a significant role.
Keywords:
Meliponine; Microsatellite markers; Males; DNA
© 2018 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
El análisis genético de congregaciones silvestres de zánganos de la abeja
sin aguijón, Scaptotrigona mexicana (Hymenoptera: Apidae) revela un elevado número de colonias en una área natural protegida del sureste de México
Resumen
En el presente trabajo se analizaron genéticamente congregaciones de zánganos de la abeja sin aguijón Scaptotrigona mexicana Guérin en 5 loci tipo microsatélite para estimar el número de colonias de esta especie en la Reserva de la Biosfera La Sepultura en Chiapas, México. En total se analizaron 216 zánganos provenientes de 3 congregaciones. Se encontró que las 3 tuvieron suficientes diferencias genéticas para ser consideradas de diferentes poblaciones El número promedio (± DE) de colonias por congregación fue de 19.8 ± 0.63, 17.6 ± 0.70 y 12; un número similar se reportó en áreas donde hay meliponicultura. Se discuten las implicaciones de estos hallazgos desde una perspectiva de conservación en la que la meliponicultura podría jugar un papel muy importante.
Palabras clave:
Meliponinos; Marcadores microsatélite; Machos; ADN
Introduction
The genetic diversity and distribution of populations in nature reserves and other protected areas are expected to be within the range of truly natural areas, unaffected by human activities such as agriculture, hunting, deforestation or pollution (Vačkářa et al., 2016). By comparing populations from protected areas with populations from geographically and climatically similar areas which are affected by human activities it is possible to infer how and to what extent human activity can shape and change population structures and dynamics (Gray et al., 2016). Mexico is a country both rich in terms of its biodiversity and its strong agriculture sector, with many crops depending upon insect pollination (Ashworth et al., 2009). As such a growing importance is given to stingless beekeeping or meliponiculture, the managing of colonies of stingless bees or meliponine (Apidae, Meliponini), since they provide pollination services to crops; also it is believed that they promote biodiversity conservation by the pollination of non-agricultural vegetation, indirectly supporting other animal species and thus general biodiversity (Cortopassi-Laurino et al., 2006).
Recent evidence shows that beekeeping seems to be not detrimental to stingless bees from a population genetic perspective; moreover, transport of colonies between meliponaries might introduce genetic novelty into managed populations, as it is the case of Melipona scutellaris Lepeletier (Carvalho-Zilse et al., 2009). Alves et al. (2010) found that even genetically impoverished meliponaries can be maintained for long time, mainly due to the energy input (care and handling of bees) by beekeepers. However, despite that managing of stingless bees might represent a good strategy for their protection, the degree to which wild populations are affected is unknown, even at the very basic level, i.e. whether meliponaries affect abundance of wild colonies; this is an important issue, since it is wild colonies what provides the gene pool that is needed to minimize the selection process during the creation of meliponaries.
Studies on the stingless bee Scaptotrigona mexicana Guérin have shown that it is feasible to estimate colony abundance in the area surrounding a meliponary by analyzing drone congregations formed in the meliponary, since in those congregations no males from managed colonies participate (Kraus et al., 2008; Müller et al., 2012). Individual genotypes usually are assigned to K-clusters through a maximum likelihood algorithm, which uses allele frequencies that are estimated from the sample from which the number of colonies is to be inferred (Wang, 2013). Drone congregations in stingless bees are clusters of males that stand near a colony expecting to mate a virgin queen from that colony (Sommeijer et al., 2004); this phenomenon is similar to the formation of drone congregations areas in the honey bee, Apis mellifera L. (Loper et al., 1992). Studies so far have shown that drone congregations can be genetically examined to explore relatedness and genetic diversity of local populations since such clusters are made up of individuals from several colonies (Baudry et al., 1998). Consequently, this information can be further used to indirectly infer how the landscape around meliponaries influences the abundance of wild colonies, only if the abundance of colonies in natural areas without beekeeping is known. This way it would be possible to give an assessment of the contribution of beekeepers to stingless bee conservation.
In this study, we aim to estimate the number of colonies and genetic population structure of the stingless bee S. mexicana in a natural setting like it is represented by a nature reserve by sampling drone congregations. With this information we could be able to gain insight into the impact and influence that stingless beekeeping has on the population genetics and overall prosperity of a stingless bee species when compared to previous findings in rural landscapes.
Materials and methods
Scaptotrigona mexicana males were carefully collected with entomological tubes from 3 congregations in the La Sepultura Biosphere Reserve, in Chiapas, Mexico (16º18’44” N, 93º49’44” W), on April 2009 (congregation 3: C3) and April 2013 (congregations 1 and 2: C1, C2). Each congregation consisted of dozens of males standing in a tree few centimeters away from the entrance of a colony. We collected 92, 82 and 42 individuals from congregations C1, C2 and C3 respectively. These numbers correspond to approximately 50% of the drones in each congregation. Congregations were spatially separated by at least 50 m from each other. Individuals were sacrificed with ethyl acetate, placed immediately in 96% ethanol and stored at -20 ºC until DNA extraction.
DNA was extracted following the HotSHOT method (Truett et al., 2000). All specimens were genotyped at 5 microsatellite loci: marker B124 developed for Bombus terrestris (Estoup et al., 1993), markers T3-32 and T4-171 for S. postica (Paxton et al., 1999) and Tc3-302, and Tc-487 for Trigona carbonaria (Green et al., 2001) following single locus routine PCR protocols (Kraus et al., 2008; Solórzano-Gordillo et al., 2015) to a final reaction volume of 5µL. Microsatellite fragments were separated using a semi-automated LI-COR 4200 slab-gel sequencer (6% denaturing polyacrylamide gels); fragment size was determined using the software SAGA MX (LI-COR Inc., US). Forward oligos were tagged with an M13(-29) sequence, which served as a priming site to a M13(-29) primer tagged with IRDYE800 (Schuelke, 2000). In order to estimate amplification errors or allele miss-callings 10% of specimens were subjected to reamplification at all loci. All reamplifications gave identical results.
Allelic richness and allelic frequency were estimated using FSTAT v2.9.3.2 (Goudet, 2001) and GENODIVE v2.0 (Meirmans & Van Tienderen, 2004), respectively. Allelic richness was Anova analyzed using R software (R Development Core Team, 2012). Potential genetic differentiation among congregations was calculated with GENEPOP on the web (Rousset, 2008); pairwise Fst comparisons were carried out with GENODIVE v2.0. In order to estimate the number of colonies we performed a sibship reconstruction based on the genotypes of the drones using the software COLONY v1.3 (Wang, 2004). We ran 10 times the sibship reconstruction with a different random seed number each time to estimate confidence intervals. Since we did not collect all males from the congregations, some colonies might be not detected. We estimated the number of undetected colonies by fitting the number of colonies and their respective number of members with a Poisson distribution (Chapman et al., 2003). Non-detection error, i.e. the probability of having genetically identical males from different fathers, was estimated for each congregation after Boomsma and Ratnieks (1996). To account for differences in sample size when comparing number of colonies among congregations we carried out resampling without replacement of genotypes from congregations C1 and C2 to get 5 subsamples of 42 genotypes; sibship reconstruction was run 2 times for each subset and the resulting number of colonies was compared with C3 with an Anova using R software.
Results
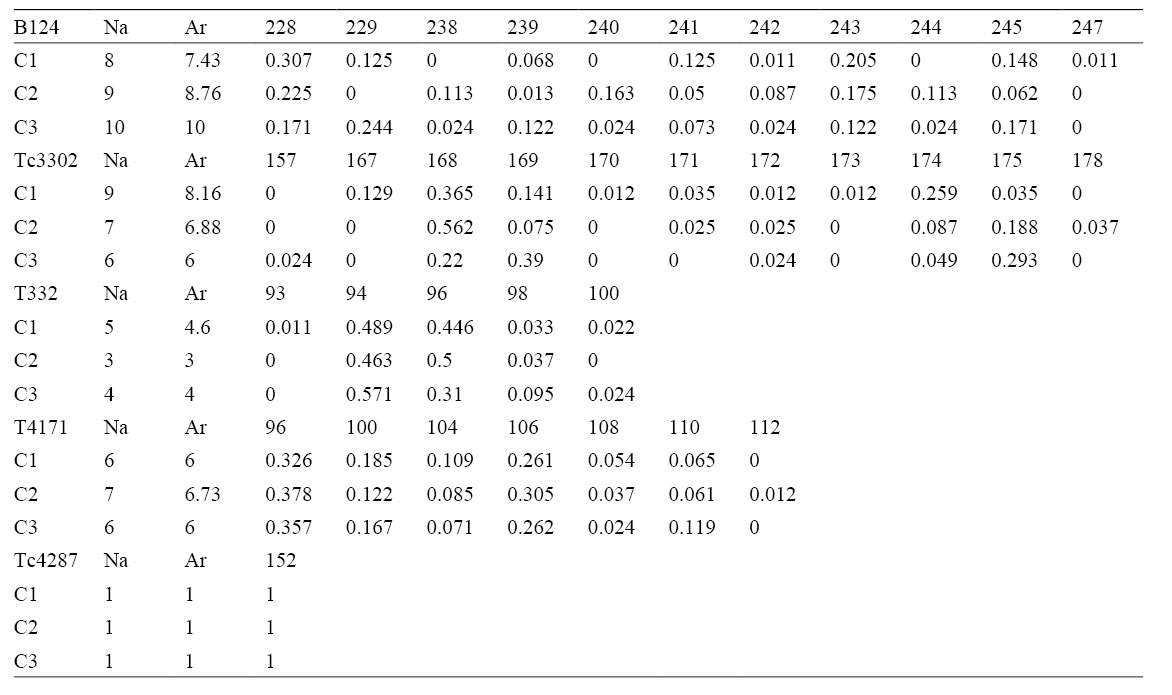
Overall 35 alleles were detected in all 3 congregations. Polymorphism varied from 1 to 11 alleles, with an overall number of 7 ± 2 (average ± SE) alleles per marker. Table 1 shows the length of fragments (in bp) and the relative frequency at each locus and congregation. Allelic richness (average ± SE) among 3 congregations (C1: 5.27 ± 1.42; C2: 5.4 ± 1.27; C3: 5.4 ± 1.47) was not significantly different (p = 0.99, F2, 12 = 0.003). Non-detection error was small in all congregations: 4.6 x 10-3, 6.6 x 10-3 and 4.6 x 10-3 for C1, C2 and C3, respectively, which did not affect our colony estimations. Congregations showed a highly significant population differentiation (Fisher’s exact test: χ2= 207.2, df = 8, p < 0.001); following pairwise tests revealed that all 3 congregations were genetically distinct (Fst’s: C1-C2 = 0.027; C1-C3 = 0.053; C2-C3 = 0.032; Amova Fst, p < 0.001 in all pairwise comparisons), thus the estimation of number of colonies was carried out separately for each congregation. The number (average ± SD) of colonies estimated was 19.8 ± 0.63, 17.6 ± 0.70 and 12 ± 0.0 for C1, C2 and C3 respectively, which gives a total of 50 colonies potentially sharing space and reproduction opportunities given the short distances between the sampling locations. The number of colonies undetected by the Poisson method was below 1 for the 3 congregations, thus we did not correct our previous estimates. Estimation of number of colonies of subsets of C1 and C2 resulted in 13.2 ± 1.31 and 13.2 ± 1.40 colonies (average ± SD), respectively. The number of colonies of C3 was not significantly different from subsets C1 and C2 at alpha = 0.01 (p = 0.03, F2, 27 = 3.9).
Discussion
In a previous work, Kraus et al. (2008) estimated that 40 and 22 wild colonies contributed the 234 Scaptotrigona mexicana males that they collected from 3 congregations in a meliponary (congregations 1 and 2 were pooled). Moreover, Müller et al. (2012) estimated that 150 drones of a single S. mexicana congregation originated from 55 colonies, again in a meliponary. In the above 2 studies, the total number of colonies was similar to our estimate of 50 colonies from 216 males collected in a nature reserve; although there is a time gap of 4 years between C1/C2 and C3, it has been shown that the average life span of stingless bees in conserved areas is over 10 years (Slaa, 2006), thus pooling the colonies from our 3 congregations is biologically plausible. Moreover, Müller et al. (2012) found that none of the colonies from the meliponary contributed to the congregations, supposedly to promote outbreeding, so a complete estimation of the actual number of colonies must include the ones in the meliponary. When we add the number of managed colonies in the meliponary to the number of wild colonies estimated by Müller et al. (2012) the number rises to 81 colonies. Though Kraus et al. (2008) did not document the number of colonies in the meliponary where the study was conducted, it is obvious that the actual number of colonies is higher if we add the managed colonies; it seems that stingless beekeeping increases the abundance of stingless bees. Such potential to sustain a higher number of colonies can be attributed to 2 factors. First, meliponaries are located in rural areas in which people grow crops mainly for self-consumption, thus there are resources available to the bees all the year; moreover, they modify the landscape to fit the nutritional needs of his/her colonies. Therefore, some of the natural resources are kept available for wild colonies. Second, beekeepers directly feed managed colonies during harsh conditions and help them to survive attacks from
Allele length and frequency of the 5 microsatellite markers used in this study for each congregation. Na, number of alleles; Ar, allelic richness.
While the number of colonies we estimated is similar to those by Kraus et al. (2008) and Müller et al. (2012), there is one clear difference between the present and those studies. Even though the 3 congregations in the nature reserve were in a very close distance to each other (average of 50 m), they displayed a clear genetic differentiation, both spatial and temporal, which is in contrast with the 2 studies in the beekeeping influenced rural environment, where the examined populations showed a much higher degree of genetic homogenization. Given the tendency of stingless bees to propagate via colony fission and the founding of daughter colonies close to the mother nests, one would in fact expect a strong genetic sub-structuring of populations given the immobility of colonies. As such, the population in the nature reserve analyzed in this study might well be a natural form of a S. mexicana population, with strong sub-structuring on a small spatial scale. The homogeneity of S. mexicana populations influenced by managed populations and beekeeping can plausibly be explained by beekeeping activities, since here we have an increased moving of colonies by beekeepers that in turn artificially enhances the gene flow resulting in stronger genetic homogenization. The long-term effects of such genetic homogenization are however difficult to predict.
The average distance that A. mellifera drones travel from their colonies to the congregations areas is around 900 m; thus drone congregations might comprise males from colonies located within a 2.5 km2 area (Moritz et al., 2008). Given the smaller size of S. mexicana compared to honey bees, it would be safe to suppose that males of this species spread over a smaller area (Araújo et al., 2004); in the case that they travel up to 400 m, our males came from 0.5 km2, i.e. 50 hectares. Fierro et al. (2012) discovered that in a landscape similar to that in which Kraus et al. (2008) collected drones, the density of wild colonies of S. mexicana was roughly one per hectare, which is in the range (0.15-15 colonies per hectare) of natural populations of many stingless bee species according to Roubik (2006). We found approximately one colony per hectare in the nature reserve, similar to Fierro et al. (2012) and 1.7 col/ha found by Slaa (2006) for another species of Scaptotrigona, S. pectoralis, in a forest reserve, which might mean that human-modified rural areas in this part of Mexico might not be as detrimental as previously thought. However, a detailed landscape analysis is necessary before making any further conjecture (Solórzano-Gordillo et al., 2015).
We could show that landscapes around meliponaries can harbor wild S. mexicana populations with carrying capacities similar to those of nature reserves, and that meliponaries themselves represent a potential refuge for stingless bees. Furthermore, the examined 3 drone congregations from a wild population exhibited genetic substructuring on a small spatial scale, probably reflecting the high genetic substructure within natural populations of that species. Our findings originate new interesting questions regarding the influence of beekeeping on the population dynamics of stingless bees: will single mating still happen at such high diversity of drones? Are other bee species, both solitary and social, positively or negatively affected by stingless beekeeping given all the changes that humans inflict on the landscape to keep economically important meliponine species?
Acknowledgements
References
Araújo, E. D., Costa, M., Chaud-Netto, J., & Fowler, H. G. (2004). Body size and flight distance in stingless bees (Hymenoptera: Meliponini): inference of flight range and possible ecological implications. Brazilian Journal of Biology, 64, 563–568.
Ashworth, L., Quesada, M., Casas, A., Aguilar, R., & Oyama, K. (2009). Pollinator-dependent food production in Mexico. Biological Conservation, 142, 1050–1057.
Baudry, E., Solignac, M., Garnery, L., Gries, M., Cornuet, J., & Koeniger, N. (1998). Relatedness among honeybees (Apis mellifera) of a drone congregation. Proceedings of the Royal Society of London B: Biological Sciences, 265, 2009–2014.
Boomsma, J. J., & Ratnieks, F. L. W. (1996). Paternity in eusocial Hymenoptera. Philosophical Transactions of the Royal Society B: Biological Sciences, 351, 947–975.
Carvalho-Zilse, G. A., Costa-Pinto, M. F., Nunes-Silva, C. G., & Kerr, W. E. (2009). Does beekeeping reduce genetic variability in Melipona scutellaris (Apidae, Meliponini)? Genetics and Molecular Research, 8, 758–765.
Chapman, R. E., Wang, J., & Bourke, A. F. G. (2003). Genetic analysis of spatial foraging patterns and resource sharing in bumble bee pollinators. Molecular Ecology, 12, 2801–2808.
Cortopassi-Laurino, M., Imperatriz-Fonseca, V. L., Roubik, D. W., Dollin, A., Heard, T., Aguilar, I. et al. (2006). Global meliponiculture: challenges and opportunities. Apidologie, 37, 275–292.
Estoup, A., Solignac, M., Harry, M., & Cornuet, J. M. (1993). Characterization of (GT)n and (CT)n Microsatellites in 2 Insect Species – Apis mellifera and Bombus terrestris. Nucleic Acids Research, 21, 1427–1431.
Fierro, M., Cruz-López, L., Sánchez, D., Villanueva-Gutiérrez, R., & Vandame, R. (2012). Effect of biotic factors on the spatial distribution of stingless bees (Hymenoptera: Apidae, Meliponini) in fragmented neotropical habitats. Neotropical Entomology, 41, 95–104.
Goudet, J. (2001). FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3.2). Available from http://www2.unil.ch/popgen/softwares/fstat.htm
Gray, C. L., Hill, S. L., Newbold, T., Hudson, L. N., Börger, L., Contu, S. et al. (2016). Local biodiversity is higher inside than outside terrestrial protected areas worldwide. Nature Communications, 7, 12306. DOI: 10.1038/ncomms12306.
Green, C. L., Franck, P., & Oldroyd, B. P. (2001). Characterization of microsatellite loci for Trigona carbonaria, a stingless bee endemic to Australia. Molecular Ecology Notes, 1, 89–92.
Kraus, F., Weinhold, S., & Moritz, R. (2008). Genetic structure of drone congregations of the stingless bee Scaptotrigona mexicana. Insectes Sociaux, 55, 22–27.
Loper, G. M., Wolf, W. W., & Taylor, Jr. O. R. (1992). Honey bee drone flyways and congregation areas: radar observations. Journal of the Kansas Entomological Society, 65, 223–230.
Meirmans, P. G., & Van Tienderen, P. H. (2004). GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Molecular Ecology, 4, 792–794.
Moritz, R. F. A., Dietermann, V., & Crewe, R. M. (2008). Determining colony densities in wild honeybee populations (Apis mellifera) with linked microsatellite DNA markers. Journal of Insect Conservation, 12, 455–459.
Müller, M. Y., Moritz, R. F. A., & Kraus, F. B. (2012). Outbreeding and lack of temporal genetic structure in a drone congregation of the neotropical stingless bee Scaptotrigona mexicana. Ecology and Evolution, 2, 1304–1311.
Paxton, R. J., Weissschuh, N., & Quezada-Euan, J. J. G. (1999). Characterization of dinucleotide microsatellite loci for stingless bees. Molecular Ecology, 8, 690–692.
R Development Core Team. (2012). R: a language and environment for statistical computing computer program. Version By R Development Core Team, Vienna, Austria.
Roubik, D. W. (2006). Stingless bee nesting biology. Apidologie, 37, 124–143.
Rousset, F. (2008). Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Molecular Ecology Resources, 8, 103–106.
Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology, 18, 233–234.
Slaa, E. J. (2006). Population dynamics of a stingless bee community in the seasonal dry lowlands of Costa Rica. Insectes Sociaux, 53, 70–79.
Solórzano-Gordillo, E., Cabrera-Marín, N. V., Mérida, J., Vandame, R., & Sánchez, D. (2015). Genetic diversity of two stingless bees, Trigona nigerrima (Cresson 1878) and Trigona corvina (Cockerell 1913), in coffee dominated landscapes in Southern Mexico. Acta Zoológica Mexicana ns, 31, 74–79.
Sommeijer, M., De Bruijn, L., & Meeuwsen, F. (2004). Behaviour of males, gynes and workers at drone congregation sites of the stingless bee Melipona favosa (Apidae: Meliponini). Entomologische Berichten, 64, 10–15.
Truett, G., Heeger, P., Mynatt, R., Truett, A., Walker, J., & Warman, M. (2000). Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques, 29, 52–54.
Vačkářa, D., Harmáčková, Z. V., Kaňková, H., & Stupková, K. (2016). Human transformation of ecosystems: comparing protected and unprotected areas with natural baselines. Ecological Indicators, 66, 321–328.
Wang, J. (2004). Sibship reconstruction from genetic data with typing errors. Genetics, 166, 1963–1979.
Wang, J. (2013). An improvement on the maximum likelihood reconstruction of pedigrees from marker data. Heredity, 111, 165–174.