Omar Lagunas-Calvo a, b, Serapio López-Jiménez c, Alejandro Oceguera-Figueroa b, *
a Universidad Nacional Autónoma de México, Posgrado en Ciencias Biológicas, Instituto de Biología, Avenida Universidad 3000, Ciudad Universitaria, 04510 Ciudad de México, Mexico
b Universidad Nacional Autónoma de México, Instituto de Biología, Departamento de Zoología, Laboratorio de Helmintología, Apartado postal 70-153, 04510 Ciudad de México, Mexico
c Universidad Juárez Autónoma de Tabasco, División Académica de Ciencias Agropecuarias, Carretera Villahermosa-Teapa Km. 25+2, Ranchería la Huasteca 2da. Sección, 86298 Villahermosa Centro, Tabasco, Mexico
*Corresponding author: aoceguera@ib.unam.mx (A. Oceguera-Figueroa)
Received: 20 July 2020; accepted: 1 March 2021
Abstract
Species included in the genus Dolops (Ichthyostraca: Argulidae) have been recorded from Southern and Central Africa, Australia, and mainly from South America, with no records from Central or North America. Specimens of Dolops bidentata, previously recorded only in South America, were collected in the state of Tabasco, southern Mexico, parasitizing the common snook, Centropomus undecimalis. Here, we provide the first record of the genus and the species in North America (Mexico) and the first molecular characterization of Dolops bidentata including 1 mitochondrial and 2 nuclear DNA markers, as well as a morphological description of the specimens. The newly generated molecular data were used to preliminarily investigate the phylogenetic relationships of Branchiura and to include Dolops bidentata in a phylogenetic hypothesis. Our results fail to recover the monophyly of Dolops; however, more investigations are needed before any taxonomic change is made.
Keywords: Parasitic crustaceans; Branchiura; Argulus; Centropomus undecimalis; COI; 18S rRNA; 28S rRNA
© 2021 Universidad Nacional Autónoma de México, Instituto de Biología. This is an open access article under the CC BY-NC-ND license
Posición filogenética de Dolops bidentata (Ichthyostraca: Argulidae) con base en datos moleculares: primer registro del género en México
Resumen
Las especies incluidas en el género Dolops (Ichthyostraca: Argulidae) han sido registradas en el sur y centro de África, Australia y principalmente en América del Sur, sin registros de América Central o del Norte. Ejemplares de Dolops bidentata, previamente registrados solo en América del Sur, se recolectaron en el estado de Tabasco, al sur de México, parasitando al róbalo Centropomus undecimalis. En el presente trabajo proporcionamos el primer registro del género y de la especie en América del Norte (México) y proporcionamos la primera caracterización molecular de Dolops bidentata que incluye un marcador de ADN mitocondrial y 2 nucleares, así como una descripción morfológica de los especímenes. La información molecular recién generada se utilizó para investigar, preliminarmente, las relaciones filogenéticas de Branchiura y para ubicar a Dolops bidentata en una hipótesis filogenética. Nuestros resultados no logran recuperar la monofilia de Dolops, sin embargo, se necesitan más investigaciones antes de realizar cualquier cambio taxonómico.
Palabras clave: Crustáceos parásitos; Branchiura; Argulus; Centropomus undecimalis; COI; 18S rARN; 28S rARN
© 2021 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
Introduction
Branchiura Thorell, 1864, commonly known as fish lice, is a group of ectoparasitic crustaceans mainly associated with freshwater and marine fishes, and less frequently amphibians (Poly, 2008). This group has a worldwide distribution, with the highest species diversity in freshwater habitats in the Afrotropical and Neotropical regions (Fryer, 1964, 1968). Currently, about 140 species arranged in 4 genera are included in the group: Argulus Müller, 1785; Chonopeltis Thiele, 1900; Dipteropeltis Calman, 1912, and Dolops Audouin, 1837 (Neethling & Avenant-Oldewage, 2016). Argulus has a cosmopolitan distribution and includes around 121 species, Chonopeltis has an Afrotropical distribution and includes 13 species, Dipteropeltis is restricted to the Neotropics and includes only 2 species, and Dolops has a Gondwanan distribution and 13 species, 11 of them restricted to the Neotropics (Fryer, 1968; Lagunas-Calvo et al., 2020; Neethling & Avenant-Oldewage, 2016; Poly, 2008).
Species included in Dolops have been recorded from Southern and Central Africa, Australia, and mainly South America (see Neethling and Avenant-Oldewage [2016] for host and locality records). To date, Dolops geayi (Bouvier, 1897), parasite of Andinoacara pulcheri (Gill, 1858), Crenicichla geayi Pellegrin, 1903, and Hoplias malabaricus (Bloch, 1794) from Lake Valencia, Venezuela, represents the northern record in America (Pearse, 1920).
Recently, fish lice specimens assigned to the genus Dolops were collected in Tabasco, southern Mexico, representing the first record for the genus in Mexico and North America. We provide the morphological description of the specimens and furthermore, we generated partial DNA sequences of both mitochondrial and nuclear genes, to characterize the species for the first time on a molecular basis and to preliminarily investigate the phylogenetic relationships of the group.
Materials and methods
In October 2019, freshwater and estuarine fish specimens of Centropomus undecimalis (Bloch, 1792), Atractosteus tropicus (Gill, 1863), Ictalurus meridionalis (Günther, 1864), and Oreochromis sp., captured by local fishermen at “El Chichicastle”, San Antonio, Pantanos de Centla, Tabasco (18°14’36.2” N, 92°18’06.5” W), were examined for ectoparasites, with special attention to the skin and gills. Fish lice specimens recovered were fixed in alcohol 80% and transported to the laboratory for further studies.
For morphological studies and measurements, 3 specimens were mounted in temporal slides and observed under a microscope. Microphotographs were taken with a Leica camera (model DFC490) attached to a Leica Z16 APO-A microscope at the Laboratorio Nacional de la Biodiversidad (LANABIO), Instituto de Biología, Universidad Nacional Autónoma de México (IBUNAM). For scanning electron microscopy (SEM) procedures, 1 specimen was dehydrated in a gradual series of ethanol from 80% to absolute, CO2 dried, covered with a Gold-Palladium mixture and observed in a Hitachi SUI510 microscope at LANABIO. A single fish lice specimen was deposited at Colección Nacional de Crustáceos (CNCR-35725), IBUNAM. Taxonomic identification was accomplished following Ringuelet (1948) and Bouvier (1899a, b).
Total DNA extraction of a single specimen was carried out using Chelex 100 (Bio-Rad) following the protocol provided by the manufacturer. Polymerase chain reactions (PCR) used 9.5 μl of DNase-free H2O, 3 μl of 5× Reaction Buffer, 0.2 μl of each primer, 0.1 μl of My Taq DNA polymerase (Bioline catalogue number BIO-21105), and 2 μl of DNA template (total volume 15 μl) for each sample. For PCR reactions with primer cocktail, the amount of DNase-free H2O was adjusted to reach a total volume of 15 μl. Mitochondrial cytochrome c oxidase subunit I (COI) DNA sequence was obtained using a primer cocktail with forward NemF1_t1+NemF2_t1+ NemF3_t1 and reverse NemR1_t1+NemR2_t1+ NemR3_t1; including 0.2 μl of each primer. PCR conditions were 94 °C for 2 min, 5 cycles at 94 °C for 40 sec, 45 °C for 40 sec, 72 °C for 1 min, followed by 35 cycles at 94 °C for 40 sec, 51 °C for 40 sec, 72 °C for 1 min, and a final extension at 72 °C for 7 min (modified from Prosser et al. [2013]). For the nuclear 18S ribosomal RNA (18S rRNA) PCR reactions, the primer cocktail included 1f+18SA2+5R+9R. PCR reactions were performed with initial denaturation at 95 ºC for 2 min followed by 35 cycles with 94 ºC for 1 min, 56 ºC for 45 sec, and 72 ºC for 45 sec, with an additional 10 min extension at 72 ºC. For the nuclear 28S ribosomal RNA (28S rRNA) amplification the primers 28sA+28SBout were used. PCR was performed with initial denaturation at 94 ºC for 5 min, followed by 35 cycles of 95 ºC for 1 min, 52 ºC for 1 min, 70 ºC for 1 min, and final extension at 72 ºC for 7 min (modified from Phillips et al. [2010]). Primer names and sequences are listed in Table 1. Amplification products were purified using CentriSep (Thermo Fisher Scientific) 96 filter plates with Sephadex G-50. Sequencing reactions used 0.4 µl BigDye Terminator ver. 3.1 (Applied Biosystems), 2 µl Buffer 5x, 4 µl ddH2O, 1 µl of primer at 10 µM, and 3 µl purified PCR product (total volume 10 µl). Primers M13F and M13R were used for sequencing the COI fragment while 18SintL and 18SintR were used for 18S. Samples were purified using Sephadex G-50, then 25 µl of 0.5 mM EDTA was added to each sample and finally sequenced in an ABI-PRISM 3100 Applied Biosystems® sequencer at LANABIO. Sequences were reconciled and edited in Geneious ver. 5.1.7 (Biomatters Ltd. Auckland, New Zealand).
The newly generated sequences were compared with the existing sequences available in the non-redundant database of the National Center for Biotechnology Information (NCBI; http://blast.ncbi.nlm.nih.gov/Blast.cgi) using the nucleotide Basic Local Alignment Search Tool (BLASTn; Altschul et al., 1990) to in order to discard the amplification of host DNA.
DNA sequences available in GenBank were selected for the phylogenetic analyses based on BLASTn results and on published literature. Two pentastomids: Levisunguis subaequalis Curran, Overstreet, Collins and Benz, 2014 and Sebekia purdieae Riley, Spratt and Winch, 1990, were selected as the outgroup based on a previous phylogenetic hypothesis (Zrzavý et al., 1997; Table 2).
The newly generated sequences were aligned together with sequences obtained from GenBank using MAFFT version 7 implemented in the web-version at https://mafft.cbrc.jp/alignment/server/ using the default parameters (Kato et al., 2019). Three matrices were assembled, 1 for each molecular marker. For the COI locus, the final matrix included 17 terminals and 675 aligned characters; this matrix was translated into amino acids using Mesquite ver. 3.61 (Maddison & Maddison, 2007) and included 224 positions; the 18S matrix included 13 terminals and 2085 aligned characters; finally, the 28S dataset included 8 terminals and 821 aligned characters. Phylogenetic analyses were performed for the COI matrix including all characters, then excluding all third positions, and finally, for the amino acid dataset in order to evaluate the occurrence of potential artifacts caused by saturation. Alternatively, based on the results of the analysis of the complete dataset and given an anomalous behavior detected in the grouping of Dolops ranarum and Argulus monodi, 2 phylogenetic analyses were conducted removing alternatively each of these taxa (Siddall & Whiting, 1999). Analyses of a concatenated dataset is precluded given the lack of common information for some terminals, for example, only outgroup taxa and D. bidentata (obtained in this study) have all 3 genetic markers available. In contrast, A. americanus, A. japonicus, and A. siamensis have no information in common.
Phylogenetic analyses were accomplished under the maximum likelihood (ML) criterion in the software RAxMLGUI ver. 2 (Silvestro & Michalak, 2011). For each DNA matrix, the GTR+GAMMA model was implemented as identified by jModelTest ver. 2 (Darriba et al., 2012) and for the analysis of the amino acid dataset, the BLOSUM62 model was implemented as identified by PartitionFinder (Lanfear et al., 2012). Maximum likelihood analyses consisted in 100 replicates using the ML + thorough bootstrap option. Bootstrap branch support values were estimated running 1,000 pseudoreplicates in the same software. The resulting tree was visualized with the software FigTree ver. 1.4.2 (Rambaut, 2012).
Results
In total, 9 specimens of C. undecimalis, 3 A. tropicus, 6 I. meridionalis, and 14 Oreochromis sp. collected in Pantanos de Centla, Tabasco, were examined. Only C. undecimalis resulted positive for ectoparasites. In total 3 specimens were recovered, 1 male and 2 females. Ectoparasites were found near to the operculum of their host.
Class Ichthyostraca Zrzavý, Hypša and Vlášková, 1997
Subclass Branchiura Thorell, 1864
Order Arguloida Yamaguti, 1963
Family Argulidae Leach, 1819
Dolops Audouin, 1837
Dolops bidentata (Bouvier, 1899a) Bouvier, 1899b
Gyropeltis bidentata Bouvier, 1899a: p. 40; Bouvier, 1899b: p. 63, figs. 2-5; Wilson, 1902: p. 736: plate XXVII: fig. 88; Malta, 1982: p. 524: figs. 4-5; 1984: p. 356; Silva-Souza et al., 2011: p. 147: figs. 1-3; Fontana et al., 2012: p. 655.
Table 1
List of primers used to obtain the DNA sequences for Dolops bidentata.
|
Gene |
Primer |
Primer sequence |
Reference |
|
Mitochondrial |
|||
|
COI |
|||
|
|
NemF1_t1 |
5’ tgtaaaacgacggccagtCRACWGTWAATCAYAARAATATTGG 3’ |
Prosser et al. (2013) |
|
|
NemF2_t1 |
5’ tgtaaaacgacggccagtARAGATCTAATCATAAAGATATYGG 3’ |
Prosser et al. (2013) |
|
|
NemF3_t1 |
5’ tgtaaaacgacggccagtARAGTTCTAATCATAARGATATTGG 3’ |
Prosser et al. (2013) |
|
|
NemR1_t1 |
5’ caggaaacagctatgactAAACTTCWGGRTGACCAAAAAATCA 3’ |
Prosser et al. (2013) |
|
|
NemR2_t1 |
5’ caggaaacagctatgactAWACYTCWGGRTGMCCAAAAAAYCA 3’ |
Prosser et al. (2013) |
|
|
NemR3_t1 |
5’caggaaacagctatgactAAACCTCWGGATGACCAAAAAATCA3’ |
Prosser et al. (2013) |
|
|
M13F |
5’ TGTAAAACGACGGCCAGT 3’ |
Messing (1993) |
|
|
M13R |
5’ CAGGAAACAGCTATGAC 3’ |
Messing (1993) |
|
Nuclear |
|||
|
18S rDNA |
|||
|
|
1f |
5’ TACCTGGTTGATCCTGCCAGTAG 3’ |
Møller et al. (2008) |
|
|
18SA2 |
5’ ATGGTTGCAAAGCTGAAAC 3’ |
Tautz et al. (1988) |
|
|
5R |
5’ CTTGGCAAATGCTTTCGC 3’ |
Møller et al. (2008) |
|
|
9R |
5’ GATCCTTCCGCAGGTTCACCTAC 3’ |
Møller et al. (2008) |
|
|
18SintR |
5’ GCG GTT AAA AAG CTC GTAG 3’ |
Møller et al. (2008) |
|
|
18SintL |
5’ TGCAACCATACTTCCCCCGG 3’ |
Møller et al. (2008) |
|
28S rDNA |
|||
|
|
28sA |
5’GACCCGTCTTGAAACACGGA3’ |
Whiting (2002) |
|
|
28SBout |
5’CCCACAGCGCCAGTTCTGCTTACC3’ |
Hovmöller et al. (2002) |
Table 2
Parasite taxa, GenBank accession number and hosts, localities, and references included in phylogenetic analyses presented herein.
|
Parasite species |
GenBank accession number |
Host (Family) |
Locality |
References |
||
|
COI |
18S rRNA |
28S rRNA |
||||
|
Pentastomida |
||||||
|
Levisunguis subaequalis |
MN062095 |
MN065568 |
MN065508 |
Gambusia affinis (Poeciliidae) |
Birmingham, Alabama, USA |
Woodyard et al. (2019) |
|
Sebekia purdieae |
KU975386 |
KU975377 |
KU975381 |
Lates calcarifer (Latidae) |
Cleveland Bay, Queensland, Australia |
Barton and Morgan (2016) |
|
Branchiura |
||||||
|
Argulus americanus |
AY456187 |
—- |
—- |
Not given |
Not given |
Lavrov et al. (2004) |
|
—- |
—- |
MN688128 |
Acipenser oxyrinchus (Acipenseridae) |
Pascagoula River, Mississippi, USA |
Andres et al. (2019) |
|
|
Argulus bicolor |
—- |
—- |
MN688129 |
Acipenser oxyrinchus (Acipenseridae) |
Pascagoula River, Mississippi, USA |
Andres et al. (2019) |
|
Table 2. Continued |
||||||
|
Parasite species |
GenBank accession number |
Host (Family) |
Locality |
References |
||
|
COI |
18S rRNA |
28S rRNA |
||||
|
Argulus bengalensis |
—- |
KM016968
|
—- |
Labeo rohita (Cyprinidae) |
Bongaon, West Bengal, India |
Patra et al. (2016) |
|
—- |
KM016969
|
—- |
Naihati, West Bengal, India |
|||
|
—- |
KF583878 |
—- |
Chakgaria, West Bengal, India |
|||
|
Argulus flavescens |
—- |
—- |
MN688125 |
Acipenser oxyrinchus (Acipenseridae) |
Pascagoula River, Mississippi, USA |
Andres et al. (2019) |
|
Argulus foliaceus |
KF713319 |
—- |
—- |
Carassius auratus (Cyprinidae) |
India |
Feroz-Khan et al. (2014) |
|
—- |
KF747861 |
—- |
Not given |
Not given |
Direct Submission |
|
|
Argulus indicus |
KF713306 |
—- |
—- |
Carassius auratus (Cyprinidae) |
India |
Feroz-Khan et al. (2014) |
|
|
KF723417 |
—- |
—- |
Not given |
Not given |
Direct Submission |
|
Argulus japonicus |
KF713304 |
—- |
—- |
Carassius auratus (Cyprinidae) |
India |
Feroz-Khan et al. (2014) |
|
KF713314 |
—- |
—- |
||||
|
KF713321 |
—- |
—- |
||||
|
GU937865 |
—- |
—- |
Not given |
Rivers, Lattakia, Syria |
Wadeh et al. (2010) |
|
|
GU937867 |
—- |
—- |
Not given |
Rivers, Guangzhou, China |
Wadeh et al. (2010) |
|
|
GU937869 |
—- |
—- |
Not given |
Rivers, Cairo, Egypt |
Wadeh et al. (2010) |
|
|
—- |
KF747860 |
—- |
Not given |
Not given |
Direct Submission |
|
|
—- |
—- |
KF747847 |
Not given |
Not given |
Direct Submission |
|
|
Argulus nobilis |
—- |
M27187 |
—- |
Lepisosteus osseus (Lepisosteidae) |
North America |
Abele et al. (1989) |
|
Argulus monody |
—- |
DQ813452 |
—- |
Heterobrachus longifilis (Clariidae) |
Lake Victoria, Tanzania |
Mwita and Nkwengulila (2010) |
|
Argulus rhipidiophorus |
—- |
KF747862 |
—- |
Not given |
Not given |
Direct Submission |
|
Argulus siamensis |
KF713308 |
—- |
—- |
Carassius auratus (Cyprinidae) |
India |
Feroz-Khan et al. (2014) |
|
KF713315 |
—- |
—- |
||||
|
KF713318 |
—- |
—- |
||||
|
—- |
KF583879 |
—- |
Cyprinus carpio (Cyprinidae) |
Chakgaria, West Bengal, India |
Patra et al. (2016) |
|
|
Dolops bidentata |
MT582371 |
MT583738 |
MT663304 |
Centropomus undecimalis (Centropomidae) |
Pantanos de Centla, Tabasco, Mexico |
Present study |
|
Dolops ranarum |
—- |
DQ813453 |
—- |
Heterobrachus longifilis (Clariidae) |
Lake Victoria, Tanzania |
Mwita and Nkwengulila (2010) |
|
Dolops sp. |
DQ889096 |
—- |
—- |
Not given |
Not given |
Costa et al. (2007) |
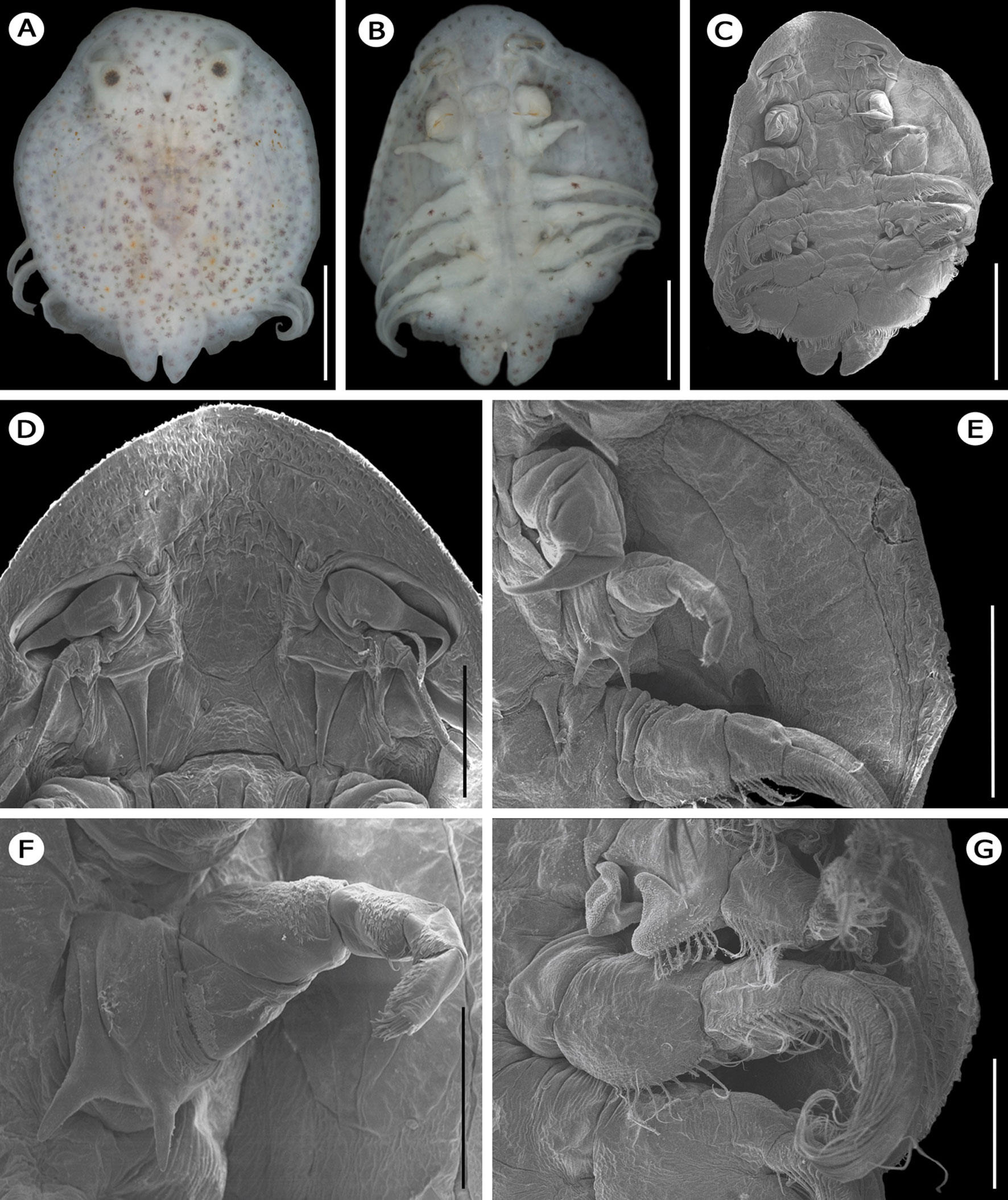
Description. Specimens are small (3.2-4.1mm length) and dorsoventrally flattened; color white-yellow. Nauplius and compound eyes present, located on dorsal surface. Carapace rounded with lateral lobes reaching the 4th pair of swimming legs. Lobes separated by a rounded sinus at the level of the 1st and 2nd legs. (Fig. 1A). Dorsal surface covered by numerous conspicuous dendriform spots, most of them brown and purple, few spots yellow (Fig. 1A). In ventral view, conspicuous dendriform spots distributed on the margins of the carapace, legs, and on natatory and abdominal lobes (Fig. 1B). Marginal spines over the entire carapace edge; extending from the anterior tip to the posterior end of the respiratory areas (Fig. 1C). Oval antero-lateral depressions, well defined (Fig. 1D). Antennules with thick rounded base and distal claws stout and tipped. Antenna thin, divided into 4 segments, with a group of bristles at the base and a group of apical spines on the terminal segment; without ornamentation at middle segments (Fig. 1D). Mouth without pre-oral sting. Suction disks absent (Fig. 1C, D). First maxillae with 2 distal claws, proximal claw smaller than the terminal (Fig. 1E). Second maxillae divided into 6 segments; first segment with 2 teeth, second segment triangular, segments 4 to 6 with pectinate scales, terminal segment with stout setae (Fig. 1E, F). Thorax formed by 4 segments, each with 1 pair of biramous legs (Fig. 1C). In males, first segment of second leg with 2 triangular projections (Fig. 1C, G). Two fusiform respiratory areas on each side of carapace. Internal respiratory area about half of the size of the external respiratory areas, in close contact to each other (1E).
Based on the lack of suction discs, second maxilla with 2 teeth at the base, and the absence of pre-oral sting, the newly collected specimens were assigned to Dolops bidentata (Bouvier, 1899). This record represents the first for the genus Dolops in Mexico and North America.
Taxonomic summary
Hosts: Anguilla sp. (Bouvier, 1899a); Astronotus ocellatus, Prochilodus nigricans, Rhytiodus microlepis, and Piaractus brachypomus (Malta, 1982. 1984); Pygocentrus nattereri (Silva-Souza et al., 2011); Schizodon fasciatus, Serrasalmus maculatus, and Serrasalmus marginatus (Fontana et al., 2012); Centropomus undecimalis (present study).
Distribution: French Guyana: Riverie Lummier (Bouvier, 1899a); Brazil: Lago Januacá, Rio Solimões, B (Malta, 1982, 1984); Coqueiro Bay (16º15’12” S, 56º22’12” W), Pirizal district, Poconé Wetland, State of Mato Grosso; and Mexico: El Chichicastle, San Antonio, Pantanos de Centla, Tabasco (18°14’36.2” N, 92°18’06.5” W at sea level).
Phylogenetic results
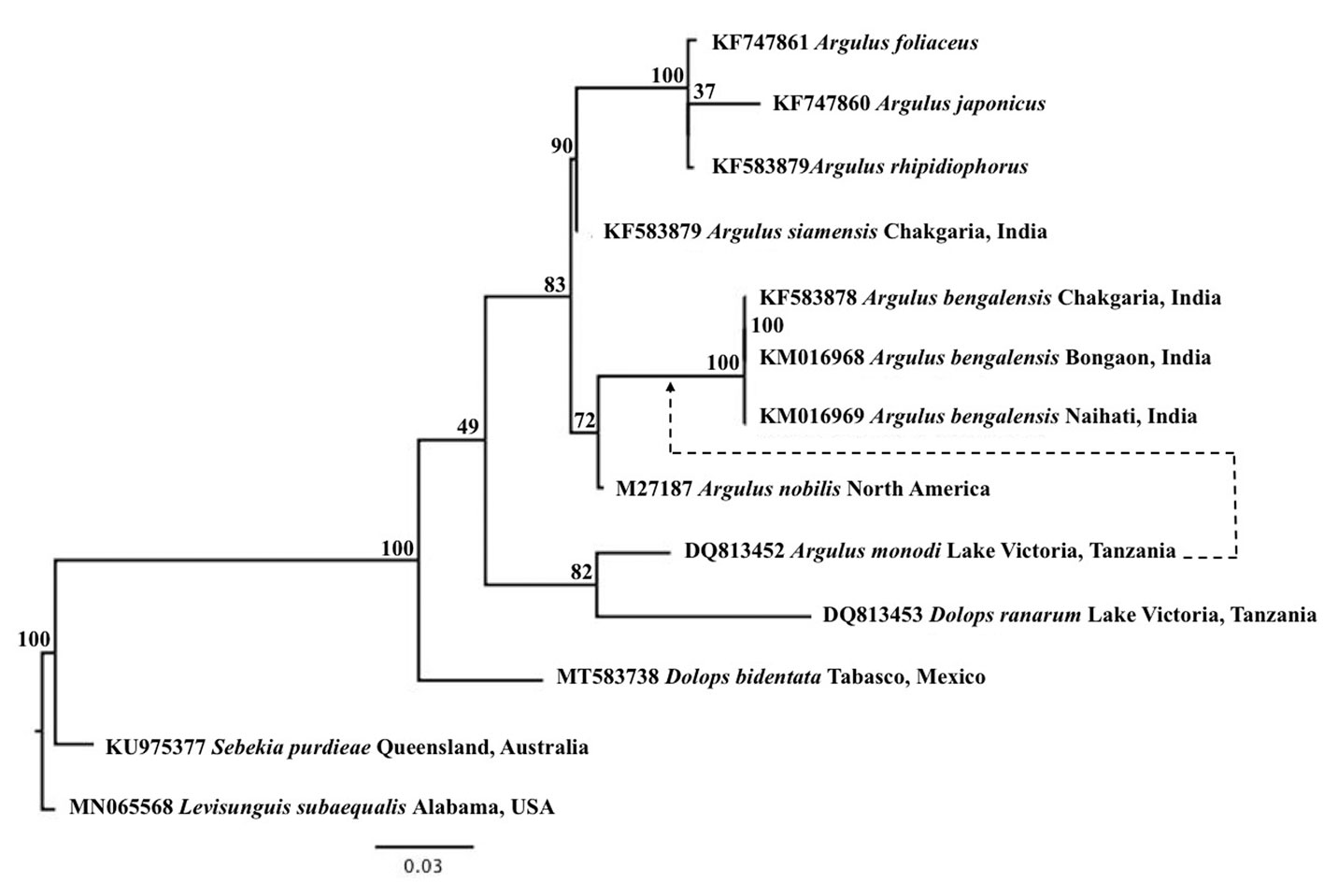
All phylogenetic trees resulting from the analyses of COI (nucleotides including and excluding third positions, and amino acids), 18S, and 28S rRNA did not recover Dolops as monophyletic. All COI trees recovered Argulus as monophyletic; Dolops sp. appears as the sister taxon of Argulus spp. and then Dolops bidentata as sister to all samples of Argulidae (Figs. 2, 3). The 18S analysis recovered Dolops as polyphyletic while Argulus was found to be paraphyletic. In this tree, D. ranarum and A. monodi, both from Lake Victoria, Tanzania, are sister taxa and show relatively long branches, while D. bidentata appears as the sister group of all samples of Argulidae (Fig. 3). Alternative exclusion of A. monodi or D. ranarum result in changes regarding the polyphyly of Dolops. Excluding A. monodi from the analyses results in the paraphyly of Dolops and monophyly of Argulus while the exclusion of D. ranarum results in the grouping of A. monodi together with A. bengalensis well nested within Argulus. Finally, the analysis of the 28S recovered D. bidentata sister to the monophyletic Argulus.
Discussion
Even though only a limited molecular dataset of Argulidae is available for phylogenetic studies, all the analyses presented here with more than 1 representative of Dolops recover the genus as either para- or polyphyletic. However, given that only 2 DNA sequences assigned to the genus Dolops are available in GenBank, our results should be taken with caution. Our phylogenetic results for Dolops disagree with current taxonomic arrangements and phylogenetic analyses based on morphological features (Bouvier, 1899a, b; Møller et al., 2008; Ringuelet, 1948). Morphological traits used to define the genus (i.e., first maxillae segmented and armed with a distal hook and by lack of the pre-oral sting) are, based on our phylogenetic results, plesiomorphic characters that transformed to a suction disk and pre-oral sting in Argulus. The transformation of such characters was investigated in previous studies (Møller & Olesen, 2010, 2012; Møller et al., 2008) but these authors concluded that it was impossible, due the lack of molecular information for the genus Dipteropeltis, to postulate the plesiomorphic condition for these features.

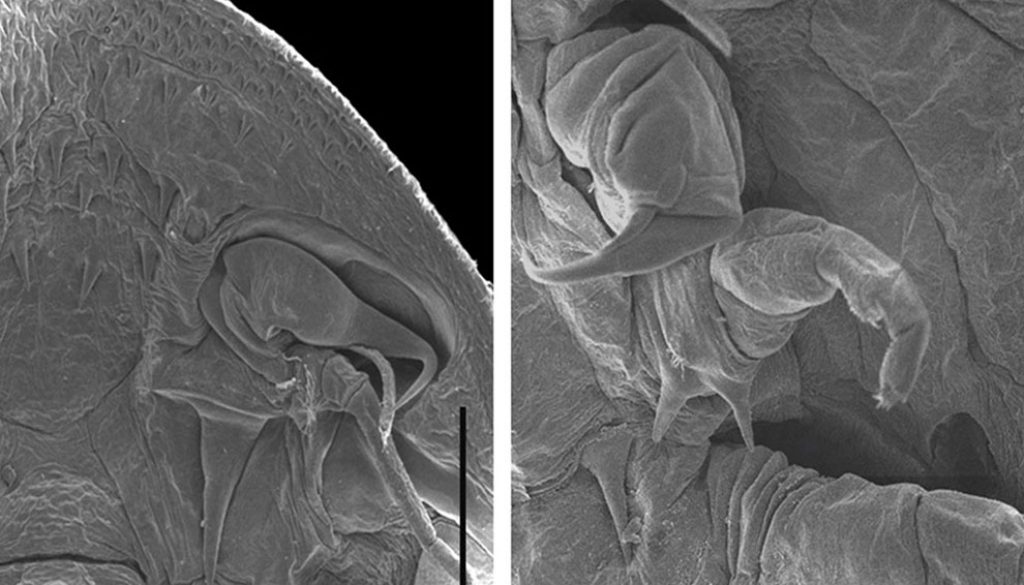
Møller et al. (2008) suggested that the presence of first maxillae segmented and armed with distal hooks, instead of suction discs, is probably the plesiomorphic condition for the family, based on the ontogenetic development (sensu Patterson [1996]). However, these authors recognized that it was equally parsimonious to assume the loss of hooks in Argulus and Chonopeltis or the loss of suction discs in Dolops. Our results suggest that the first maxillae segmented and armed with distal hooks is the plesiomorphic condition in Branchiura, based on the position of species of Dolops in all the phylogenetic analysis performed herein.
Regarding the absence of the pre-oral sting in Dolops, our results corroborate the preliminary discussion of Møller and Olesen (2010). These authors proposed that the acquisition of the pre-oral sting probably occurred in the common ancestor of Dipteropeltis, Argulus, and Chonopeltis, and this latter genus lost this character secondarily. Based on the position of species of Dolops in our trees and previous studies that show that this structure is absent in newly hatched larvae (stage 1) of D. ranarum and D. carvalhoi (Avenant-Oldewage et al., 1989; Fryer, 1964; Møller & Olesen, 2012), we propose that the lack of pre-oral sting is the plesiomorphic condition for Branchiura.
The presence of 2 teeth at the base of second maxillae is the distinctive feature of D. bidentata and is clearly described in the original description (Bouvier, 1899a), as well as in the expanded description of the species (Bouvier, 1899b). This feature is also present in the specimens studied here collected in Mexico as well as in the specimens collected in the state of Mato Grosso, Brazil, parasitizing the red piranha that were used for the redescription of the species by Silva-Souza et al. (2011). Interestingly, other than this conspicuous feature, specimens form Mato Grosso display morphological characters clearly divergent from those described in the original description and those found on the specimens from Mexico. In both the original description and in the specimens from Mexico, the dorsal surface of the body is covered by numerous dendriform spots not forming regular patterns; most of the spots are brown or purple with few spots in yellow, whereas in the specimens from Mato Grosso, spots are arranged on the borders of carapace and the external borders of abdominal lobes, lateral lines at the middle of lateral lobes, 1 on each side and lines surrounding both eyes, and furthermore, all the spots are black. In addition, the specimens from Mato Grosso lack of a nauplius eye whereas this trait is described in both the original description and in the specimens from Mexico. Furthermore, lateral lobes of the carapace in the specimens from Mato Grosso only reach the 3rd pair of swimming legs, whereas in the Mexican specimens and in the original description of the species, both lobes reach the 4th pair of swimming legs.
One of the main differences between the specimens from Mato Grosso described by Silva-Souza et al. (2011) in comparison with the specimens from Mexico is the shape, size, and arrangement of the respiratory areas. In the specimens from Mato Grosso, internal respiratory areas are small and oval and external respiratory areas large and fusiform with an indentation on the internal side in the posterior part; internal and external areas are well separated from each other. On the contrary, in Mexican specimens, internal and external respiratory areas are fusiform and in close contact to each other. Unfortunately, no information of such characters is available in the original description of the species.
In addition, differences in antennules, male accessory copulatory structures, and abdominal lobes between the original description and the specimens from Mexico in comparison with the specimens from Mato Grosso were found. In specimens described by Silva-Souza et al. (2011: p. 147: fig. 2b), each antennule has a large robust lateral spine and 4 apical spines. In contrast, these spines are not described in Bouvier (1899a, b) and are not present in our material. In both Mexican specimens and in the original description, males have 2 triangular accessory copulatory structures in the second legs, while in the material from Mato Grosso, similar modifications are observed in legs 2 to 4 (Silva-Souza et al., 2011: p: 147: figs. 2g-i). Bouvier (1899b: p. 66: fig. 5c) described 2 laminar projections at the base of third legs similar to those described by Silva-Souza et al. (2011), but such structures were not observed in our material. Abdominal lobes in the specimens from the 3 localities are similar in size, shape, and in the presence of small spines along their margin, but specimens from Mato Grosso have both lobes separated by a broad sinus about half the length of the complete abdomen, while in the Mexican specimens, the sinus is shorter, as in the description of Bouvier (1899b: p. 63: fig. 2a).
Morphologically, the material collected in Mexico resembles the original description of the species by Bouvier (1899a, b), whereas major differences were found in comparison with the specimens described by Silva-Souza et al. (2011). Based on this, we assign our specimens to D. bidentata, whereas the material from Brazil probably corresponds to a different species.
Acknowledgments
Andrea Jiménez Marín, Andrea Rebollo Hernández, Laura Márquez Valdelamar, and Nelly López Ortíz (LANABIO, UNAM) assisted in the generation of DNA sequences. Berenit Mendoza Garfias and Susana Guzmán-Gómez assisted in the microphotograph processing. This project was funded by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica IN213520 (PAPIIT-UNAM) to AO-F. Conacyt provided a scholarship to LC-O. We also thank Jose Luis Villalobos and Fernando Álvarez for incorporating the specimens into the Colección Nacional de Crustáceos, Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City. This paper is part of the first author research in the Posgrado en Ciencias Biológicas-UNAM.
References
Abele, L. G., Kim, W., & Felgenhauer, B. E. (1989). Molecular evidence for inclusion of the phylum Pentastomida in the Crustacea. Molecular Biology and Evolution, 6, 685–691. https://doi.org/10.1093/oxfordjournals.molbev.a040581
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Andres, M. J., Higgs, J. M., Grammer, P. O., & Peterson, M. S. (2019). Argulus from the Pascagoula River, MS, USA, with an emphasis on those of the threatened Gulf Sturgeon, Acipenser oxyrinchus desotoi. Diversity, 11, 232. https://doi.org/10.3390/d11120232
Avenant-Oldewage, A., van As, J. G., & Loots, G. C. (1989). On the hatching and morphology of Dolops ranarum larvae (Crustacea: Branchiura). Journal of Zoology, 217, 511–519.
Barton, D. P., & Morgan, J. A. (2016). A morphological and genetic description of pentastomid infective nymphs belonging to the family Sebekidae Sambon, 1922 in fish in Australian waters. Folia Parasitologica, 63, 1. https://doi.org/10.14411/fp.2016.026
Bouvier, E. L. (1899a). Les Crustacés parasites du genre Dolops Audouin (Première Partie). Bulletin de la Société Philomathique de Paris, 8, 53–81.
Bouvier, E. L. (1899b). Les Crustacés parasites du genre Dolops Audouin (Seconde Partie). Bulletin de la Société Philomathique de Paris, 9, 12–40.
Costa, F. O., DeWaard, J. R., Boutillier, J., Ratnasingham, S., Dooh, R. T., Hajibabaei, M. et al. (2007). Biological identifications through DNA barcodes: the case of the Crustacea. Canadian Journal of Fisheries and Aquatic Sciences, 64, 272–295. https://doi.org/10.1139/f07-008
Darriba, D., Taboada, G. L., Doallo, R, & Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9, 772. https://doi.org/10.1038/nmeth.2109
Feroz-Khan, K., Sanker, G., & Prasanna-Kumar, C. (2014). Linking eggs and adults of Argulus spp. using mitochondrial DNA barcodes. Mitochondrial DNA Part A, 27, 3927–3931. https://doi.org/10.3109/19401736.2014.987269
Fontana, M., Takemoto, R. M., Malta, J. C. O., & Mateus, L. A. F. (2012). Parasitism by argulids (Crustacea: Branchiura) in piranhas (Osteichthyes: Serrasalmidae) captured in the Caiçara bays, upper Paraguay River, Pantanal, Mato Grosso state, Brazil. Neotropical Ichthyology, 10, 653–659. https://doi.org/10.1590/S1679-62252012005000019
Fryer, G. (1964). Further studies on the parasitic Crustacea of the African freshwater fishes. Proceedings of the Zoological Society of London, 143, 79–102.
Fryer, G. (1968) The parasitic Crustacea of African freshwater fishes; their biology and distribution. Journal of Zoology,
156, 45–95. https://doi.org/10.1111/j.1469-7998.1968.tb085
78.x
Hovmöller, R., Pape, T., & Källersjö, M. (2002). The Palaeoptera problem: basal pterygote phylogeny inferred from 18S and 28S rDNA sequences. Cladistics, 18, 313–323. https://doi.org/10.1111/j.1096-0031.2002.tb00153.x
Katoh, K., Rozewicki, J., & Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics, 20, 1160–1166. https://doi.org/10.1093/bib/bbx108
Lagunas-Calvo, O., García-Prieto, L., Osorio-Sarabia, D., León-Règagnon, V., & Oceguera-Figueroa, A. (2020). New records of Ichthyostraca Zrzavý, Hypša & Vlášková, 1997 (Pancrustacea) from Mexico with an annotated checklist of North America. Zootaxa, 4775, 1–55. https://doi.org/10.11646/zootaxa.4755.1.1
Lanfear, R., Calcott, B., Ho, S. Y., & Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29, 1695–1701. https://doi.org/10.1093/molbev/mss020
Lavrov, D. V., Brown, W. M., & Boore, J. L. (2004). Phylogenetic position of the Pentastomida and (pan) crustacean relationships. Proceedings of the Royal Society of London. Series B: Biological Sciences, 271, 537–544. https://doi.org/10.1098/rspb.2003.2631
Maddison, W. P., & D. R. Maddison. (2007). Mesquite: a modular system for evolutionary analysis. Version 2.0 http://mesquiteproject.org
Malta, J. C. D. O. (1982). Os argulídeos (Crustacea: Branchiura) da Amazônia Brasileira. Aspectos da ecologia de Dolops discoidalis Bouvier, 1399 e Dolops bidentata Bouvier, 1899. Acta Amazonica, 12, 521–528. https://doi.org/10.1590/1809-43921982123521
Malta, J. C. D. O. (1984). Os peixes de um lago de Várzea da Amazônia central (Lago Januacá, Rio Solimões) e suas relações com os crustáceos ectoparsitas (Branchiura: Argulidae). Acta Amazonica, 14, 355–372. https://doi.org/10.1590/1809-43921984143372
Messing, J. (1993). M13 cloning vehicles: their contribution to DNA sequencing. Methods in Molecular Biology, 23, 9–22. https://doi.org/10.1385/0-89603-248-5:9.
Møller, O. S., & Olesen, J. (2010). The little-known Dipteropeltis hirundo Calman, 1912 (Crustacea, Branchiura): SEM investigations of paratype material in light of recent phylogenetic analyses. Experimental Parasitology, 125, 30–41. https://doi.org/10.1016/j.exppara.2009.09.008
Møller, O. S., & Olesen, J. (2012). First description of larval stage 1 from a non-African fish parasite Dolops (Branchiura). Journal of Crustacean Biology, 32, 231–238. https://doi.org/10.1163/193724011X615541
Møller, O. S., Olesen, J., Avenant-Oldewage, A., Thomsen, P. F., & Glenner, H. (2008). First maxillae suction discs in Branchiura (Crustacea): development and evolution in light of the first molecular phylogeny of Branchiura, Pentastomida, and other “Maxillopoda”. Arthropod Structure and Development, 37, 333–346. https://doi.org/10.1016/j.asd.2007.12.002
Mwita, C. J., & Nkwengulila, G. (2010). Phylogenetic relationships of the metazoan parasites of the clariid fishes of Lake Victoria inferred from partial 18S rDNA sequences. Tanzania Journal of Science, 36, 47–57.
Neethling, L. A., & Avenant-Oldewage, A. (2016). Branchiura —a compendium of the geographical distribution and a summary of their biology. Crustaceana, 89, 1243–1446. https://doi.org/10.1163/15685403-00003597
Patra, A., Mondal, A., Banerjee, S., Adikesavalu, H., Joardar, S. N., & Abraham, T. J. (2016). Molecular characterization of Argulus bengalensis and Argulus siamensis (Crustacea: Argulidae) infecting the cultured carps in West Bengal, India using 18S rRNA gene sequences. Molecular Biology Research Communications, 5, 156–166. https://doi.org/10.22099/MBRC.2016.3752
Patterson, C. (1996). Comments on Mabee’s “Empirical rejection of the ontogenetic polarity criterion”. Cladistics, 12, 147–168. https://doi.org/10.1111/j.1096-0031.1996.tb00199.x
Pearse, A. S. M. (1920). The fishes of Lake Valencia, Venezuela. University of Wisconsin Studies in Science, 12, 1–51. https://doi.org/10.5962/bhl.title.18320
Phillips, A. J., Arauco-Brown, R., Oceguera-Figueroa, A., Gomez, G. P., Beltrán, M., Lai, Y. T. et al. (2010). Tyrannobdella rex n. gen. n. sp. and the evolutionary origins of mucosal leech infestations. Plos One, 5, e10057. https://doi.org/10.1371/journal.pone.0010057
Poly, W. J. (2008). Global diversity of fishlice (Crustacea: Branchiura: Argulidae) in freshwater. Hydrobiologia, 595, 209–212. https://doi.org/10.1007/978-1-4020-8259-7_22
Prosser, S. W. J., Velarde-Aguilar, M. G., León-Règagnon, V., & Hebert, P. D. N. (2013). Advancing nematode barcoding: A primer cocktail for the cytochrome oxidase subunit I gene from vertebrate parasitic nematodes. Molecular Ecology Resources, 13, 1108–1115.
Rambaut, A. (2012). FigTree v1.4. Available from http://tree.bio.ed.ac.uk/software/figtree/
Ringuelet, R. A. (1948). Argulídos del Museo de la Plata. Revista del Museo de La Plata, 5, 281–296.
Siddall, M. E., & Whiting, M. F. (1999). Long-branch abstractions. Cladistics, 15, 9–24. https://doi.org/10.1111/
j.1096-0031.1999.tb00391.x
Silva-Souza, A. T., Abdallah, V. D., De Azevedo, R. K., Da Silva, F. A., & Luque, J. L. (2011). Expanded description of Dolops bidentata (Bouvier, 1899) (Branchiura: Argulidae) based on specimens collected on Pygocentrus nattereri Kner, 1858 (Characiformes) from Poconé Wetland, MT, Brazil. Brazilian Journal of Biology, 71, 145–149. https://doi.org/10.1590/S1519-69842011000100021
Silvestro, D., & Michalak, I. (2011). RaxmlGUI: A graphical front-end for RAxML. Organisms Diversity and Evolution, 12, 335–337. https://doi.org/10.1007/s13127-011-0056-0
Tautz, D., Hancock, J. M., Webb, D. A., Tautz, C., & Dover, G. A. (1988). Complete sequences of the rRNA genes of Drosophila melanogaster. Molecular Biology and Evolution, 5, 366–376. https://doi.org/10.1093/oxfordjournals.molbev.a040500
Wadeh, H., Alsarakibi, M., & Li, G. (2010). Analysis of genetic variability within Argulus japonicus from representatives of Africa, Middle East, and Asia revealed by sequences of three mitochondrial DNA genes. Parasitology Research, 107, 547–553. https://doi.org/10.1007/s00436-010-1891-1
Whiting, M. F. (2002). Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoological Scripta, 31, 93–104. https://doi.org/10.1046/j.0300-3256.2001.00095.x
Wilson, C. B. (1902). North American parasitic copepods of the family Argulidae, with a bibliography of the group and a systematic review of all known species. Proceedings of the United States National Museum, 25, 635–742. https://doi.org/10.5479/si.00963801.25-1302.635
Woodyard, E. T., Stilwell, J. M., Camus, A. C., & Rosser, T. G. (2019). Molecular and histopathological data on Levisunguis subaequalis Curran, Overstreet, Collins and Benz, 2014 (Pentastomida: Eupentastomida: Porocephalida: Porocephaloidea: Sebekidae: Sebekinae) from Gambusia affinis in Alabama, USA. Journal of Parasitology, 105, 827–839. https://doi.org/10.1645/19-38
Zrzavý, J., Hypša, V., & Vlášková, M. (1997). Arthropod phylogeny: taxonomic congruence, total evidence and conditional combination approaches to morphological and molecular data sets. In R. A. Fortey, & R. H. Thomas, (Eds.), Systematics Association, Special Volume Series 55. London: Chapman and Hall.