Cinthya Leocadio a, Nohely Álvarez-López b, Alejandra Barrios a, Abraham Guerra c, Yunuen Tapia-Torres b, Patricia Velez a, *
a Universidad Nacional Autónoma de México, Instituto de Biología, Tercer Circuito s/n, Coyoacán, 04510 Ciudad de México, Mexico
b Universidad Nacional Autónoma de México, Escuela Nacional de Estudios Superiores-Unidad Morelia, Antigua Carretera a Pátzcuaro No. 8701, Ex Hacienda de San José de la Huerta, 58190 Morelia, Mexico
c Universidad Simón Bolívar, Facultad de Ciencias Básicas y Biomédicas, Calle 58 #55-132, Sede 3, Barranquilla, Colombia
*Corresponding author: pvelez@ib.unam.mx (P. Velez)
Received: 18 February 2022; accepted: 27 January 2023
Abstract
Montane cloud forests are among the most threatened ecosystems globally. These forests face several stressors, such as deforestation and climate change, jeopardizing their functional sustainability. Although microbial communities act as key regulators of the soil nutrient cycles, microfungal and bacterial diversity remains largely unknown in this ecosystem. We evaluated cultivable soil microbial diversity associated with the soil below iconic plant taxa (Cyatheaceae and Juglandaceae) in a pristine montane cloud forest of Mexico, and explored small-scale ecological patterns linked to edaphic biogeochemical variables. Our findings revealed the copious occurrence of entomopathogenic fungi such as Tolypocladium geodes and potentially phosphate solubilizer bacteria such as Pseudomonas and Bacillus spp. We observed a strong association between edaphic microbial assemblages and environmental variables such as soil C:N:P availability. This close relationship with the physical setting should be considered for the development of management and in situ conservation strategies aiming to preserve microbial functions.
Keywords: Microbial edaphic diversity; C:N:P stoichiometry; Spatial heterogeneity; Phytopathogenic fungi
© 2023 Universidad Nacional Autónoma de México, Instituto de Biología. This is an open access article under the CC BY-NC-ND license
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Diversidad microbiana cultivable del suelo en un bosque mesófilo de montaña prístino en Oaxaca, México
Resumen
El bosque mesófilo de montaña se encuentra entre los ecosistemas más amenazados a escala mundial. Actualmente, enfrenta diversos disturbios de origen antrópico, tales como la deforestación y el cambio climático, que comprometen su sostenibilidad funcional. A pesar de que las comunidades microbianas fungen como reguladoras de los ciclos de nutrientes en el suelo, su diversidad permanece desconocida en gran medida para el bosque mesófilo. En el presente estudio evaluamos la diversidad y la estructura de comunidades bacterianas y fúngicas asociadas con el suelo circundante a especies vegetales icónicas (Cyatheaceae y Juglandaceae) en una localidad prístina de bosque mesófilo de montaña en México, y exploramos patrones ecológicos a una escala geográfica fina vinculados con variables biogeoquímicas edáficas. Nuestros resultados revelaron una alta prevalencia de especies de hongos entomopatógenos como Tolypocladium geodes y de bacterias potencialmente solubilizadoras de fosfato como Pseudomonas y Bacillus spp. Observamos una fuerte asociación entre la composición de las comunidades fúngicas y bacterianas con variables ambientales clave, tales como la disponibilidad de C:N:P en el suelo. Estos resultados deben ser considerados para el desarrollo y aplicación de estrategias de conservación in situ con el objetivo de preservar las funciones microbianas.
Palabras clave: Diversidad edáfica microbiana; Estequiometría C:N:P; Heterogeneidad espacial; Hongo fitopatógeno
© 2023 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
Introduction
Montane cloud forests (MCF) cover 0.26% of the Earth’s surface, and less than 1% of the Mexican territory (Bubb et al., 2004). This ecosystem is characterized by a persistent cloud immersion (Rosas Rangel et al., 2019), occurring as patches at elevations of 600-3,200 m asl (Alfonso-Corrado et al., 2017; Ochoa-Ochoa et al., 2017; Santillán et al., 2020). It hosts a number of macroscopic endemic species (12% of the overall American mammal, bird and amphibian species; Hamilton, 2009; Karger et al., 2021), being recognized for its notorious levels of fungal diversity even at the small scale (Velez et al., 2021). The MCF provides vital ecosystemic services such as carbon capture, erosion control, as well as climate regulation, soil fertility, water supply and quality (Bazzaz, 1998; Bruijnzeel et al., 2010, 2011; Martínez et al., 2009). However, this unique biome ranks among the most threatened ecosystems globally, facing several stressors such as deforestation (Leija-Loredo & Pavón, 2017), climate change (Alfonso-Corrado et al., 2017), reduction of humidity (Santillán et al., 2020), increments in temperature (Foster, 2001), among others.
Microbial communities constitute important soil components (De Long et al., 2019) that fulfill key roles in edaphic nutrient cycles, serving as a sink and source of nutrients due to their remarkable ability to immobilize and release carbon (C), nitrogen (N), and phosphorus (P) in different chemical forms (Zak et al., 2003). This group includes several taxonomic assemblies (e.g., fungi, bacteria, virus, archaea and protists) of organisms smaller than 100µm (Wagg et al., 2018). Among these taxa, bacteria and fungi (hereafter referred to as a microbial assemblage, sensu Nemergut et al., 2013) are the largest and most diverse components comprising up to 90% of the overall microbial biomass in soils (Rinnan & Bååth, 2009). Hence, the understanding of this imperceptible, yet large component of soil diversity in MCFs represents a fundamental element for conservation.
Data from microcosm and field studies have demonstrated that microbial diversity and community composition influence soil ecosystem process rates (McGuire & Treseder, 2010). In this sense, bacteria and fungi collaborate in the decomposition and mineralization of organic remains (Romaní et al., 2006; Tapia-Torres & García-Oliva, 2013), driving the development of edaphic stable and labile pools of C, N and other nutrients, which facilitate the subsequent establishment of plant communities (Schulz et al., 2013). In forest systems, bacteria carry out the hydrolysis and mineralization of organic matter through the biosynthesis of exoenzymes, followed by the release and uptake of nutrients (PO4– and NH4+) from the soil solution. Emblematic taxa with these capacities include members of Pseudomonas, Burkholderia, Escherichia, Serratia, Bacillus, Enterobacter, Nostoc, Caulobacter, Sinorhizobium, Mesorhizobium, and Corynebacterium (Horwath, 2017; Idriss et al., 2002).
Additionally, edaphic fungi perform several ecological roles as pathogens, saprotrophs, and symbionts (Nguyen et al., 2016). These osmotrophs play essential roles in nutrients turnover (Zanne et al., 2020), depolymerizing recalcitrant lignin and cellulose molecules contained in leaf and wood litter through the production of extracellular enzymes (de Boer et al., 2005). Furthermore, fungal pathogenic taxa act as biological control agents, being implicated in plant diversity maintenance (Brown et al., 2011). To the best of our knowledge, soil microfungal diversity in Mexican MCFs includes members affiliated to Alternaria, Aspergillus, Bipolaris, Chaetomium, Cladosporium, Cordana, Curvularia, Chalara, Dictyochaeta, Fusarium, Gyrothrix, Humicola, Monodictys, Myrmecridium, Penicillium, Periconia, Pestalotiopsis, Sporidesmium, Stachybotrys, Talaromyces, Trichoderma, and Virgaria (Arias & Heredia-Abarca, 2014, 2020; Heredia-Abarca et al., 2011). Also, several ectomycorrhizal fungi have been linked with roots of Juglandaceae species (Corrales et al., 2021).
The generation and amalgamation of diversity data at different scales is fundamental to develop a broad understanding of ecosystems (Oda et al., 2019). At the large scale, MCFs have been extensively investigated, reporting high heterogeneity and diversity levels (Williams et al., 2013). Though, small-scale studies have received less attention with respect to larger macroscale explorations. Pioneer efforts analyzing biogeochemical data have demonstrated an environmentally heterogeneous setting, with enzymatic activities suggesting distinctive small-scale soil patterns (Velez et al., 2021). Nevertheless, microbial diversity patterns remain poorly understood at the small scale in this environment, hampering the robust view of ecosystem functioning as small-scale processes may be masked by larger scale features (Mori et al., 2018).
In view of MCFs vulnerability to anthropogenic stressors, and the lack of knowledge on soil microbial diversity and its relationship with ecosystem processes (e.g., nutrient cycling) at different scales, herein we evaluated cultivable soil microbial diversity and community structure associated with the soil below 2 iconic plant taxa (endemic, relict, and endangered species) in a pristine location of Mexican MCF at the small spatial scale. We predict that our approach will lead to the predominant isolation of saprotrophic fungi and potentially phosphate solubilizer bacteria; in addition, we hypothesize that the small-scale distribution of microbial assemblages will be strongly associated with environmental variables such as soil phosphorous availability.
Materials and methods
The fieldwork was conducted in the MCF locality of El Relámpago (17°35’30.4” N, 96° 23’57.1” W; at 2,219 m asl), within the municipality of Santiago Comaltepec, in the mountainous system of northern Oaxaca (del Mar Delgado-Serrano et al., 2015). This forest harbors high numbers of endemic vertebrate species and a genetically diverse population of Oreomunnea mexicana, due to its good conservation status (del Mar Delgado-Serrano et al., 2015; Ponce-Reyes et al., 2012, 2020). The climate is usually temperate-humid with rainfall in summer (INEGI, 2010), an annual average temperature of 16-20 °C, and average annual precipitation of 2,000-4,500 mm (Trejo, 2004). The main soil type is Acrisol, which is strongly acidic, with a subsurface horizon of clay accumulation and low nutrient retention capacity (Alfaro-Sánchez, 2004; Krasilnikov et al., 2013). Velez et al. (2021) described the biogeochemical characteristics of the study site, highlighting high edaphic heterogeneity at the small spatial scale, abundant total carbon (TC) and dissolved organic carbon (DOC), as well as high polyphenol oxidase (POX) activity values.
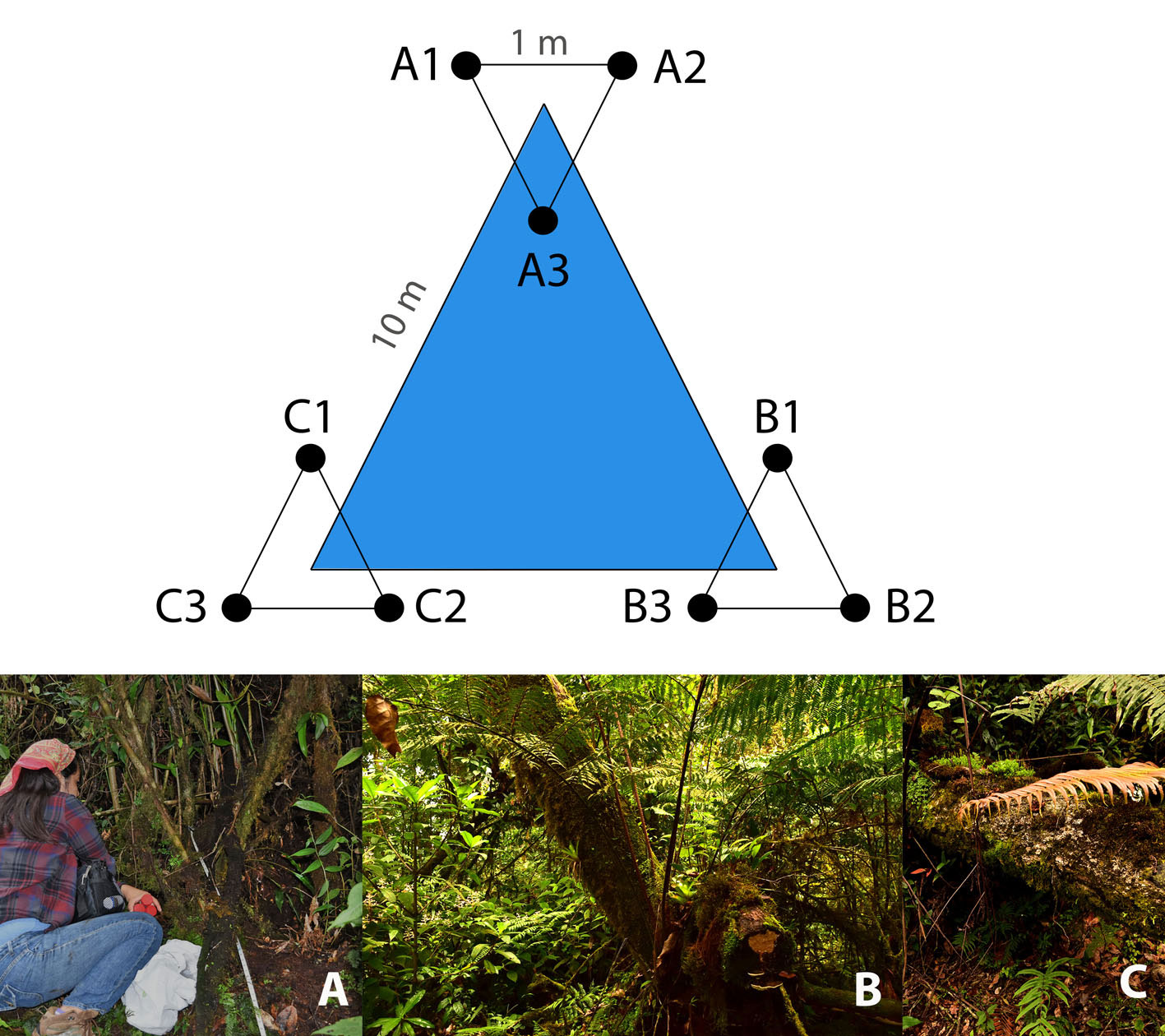
We employed a triangular sampling method as proposed by Bąk (2014). Therefore, 3 sampling plots were set up along a 10 m-triangular transect, considering different elements of MCF for a greater representation of the microbial community: the first plot was established adjacent to an individual of O. mexicana (17°35’0.36” N, 96°23’36.6” W), a representative and endangered species of Mexican MCF (Alfonso-Corrado et al., 2017; Rzedowski, 1996); the second plot was settled next to an individual of Alsophila salvinii (17°35’18.4” N, 96°23’57.7” W), a conspicuous fern in the region (Rzedowski & Palacios-Chávez, 1977); and the third plot corresponded to the area under a fallen wooden log (17°34’51.83” N, 96°24’5.17” W). Subsequently, in each plot a 1 m-equilateral triangular subplot was traced (Fig. 1). In total, 9 soil cores (3 per plot) were sampled in the first 10 cm of soil (excluding litter) using sterile Falcon tubes and transported in a cooler to the laboratory within the next 48 h for processing.
During the fieldwork we detected a visibly sick population of A. salvinii (presence of dark spots and blights on fronds), including the individual within our sampling plot (Fig. 2). So, despite that this was not part of the objectives of this study, and given the importance of prompt disease detection in threatened ecosystems in order to mitigate outbreaks, samples (consisting of sick fronds individually placed in Zip-lock® plastic bags) were collected to characterize the etiological agent. All the material was immediately stored and transported at 4º C in the dark to the laboratory and processed within the next 48 h.
Fungi and bacteria were isolated using the dilution plating method (Warcup, 1960) on Potato Dextrose Agar (PDA; Fluka Analytical, Sigma-Aldrich), Corn Meal Agar (CMA; Fluka Analytical, Sigma-Aldrich), Luria Bertani Agar (LB; Lennox Agar, Invitrogen), and LB− (2-fold diluted LB). Dilution plates were prepared using 1 g of soil sample, at 10-1 – 10-6 dilutions in test tubes with sterilized distilled water. Three replicates for each dilution of each sample were plated (1 ml aliquot). Petri dishes were incubated at laboratory room temperature (22-25 °C), with a 12 h photoperiod, and examined periodically for up to 2 weeks. During this period, different colony morphologies from each medium were transferred and maintained on PDA and LB plates for fungi and bacteria respectively.
Fronds from A. salvinii were initially washed in running tap water. Next, surface sterilization was attained by sequential solutions of 70% ethanol (1 min), 2.6% sodium hypochlorite (3 min), and 70% ethanol (1 min). Small pieces (0.5 × 0.5 cm) from sections including dark spots and blights on leaves were placed in Petri dishes containing PDA, and incubated at 25 °C for 10 to 15 days. Following incubation, fungal axenic isolates were obtained and subsequently transferred to PDA for maintenance.
Fungi were identified by evaluating morphological characteristics, combined with the analysis of the ITS1-5.8S-ITS2 rDNA, hereafter referred to as the ITS region. So, genomic DNA of the axenic isolates was extracted using the protocol described by Doyle and Doyle (1987). The ITS region was amplified by using primers set ITS1 and ITS4 as reported by White et al. (1990). The bacterial genomic DNA from axenic cultures was isolated using Dneasy Blood & Tissue Kit®. The 16S ribosomal DNA region was amplified with primers 27F and 1492R (Lane, 1991). The PCR products were sequenced in both directions using a 3730xl DNA Analyzer (Applied Biosystems™) at LANABIO, Biology Institute, National Autonomous University of Mexico (UNAM). Cultures and total DNA are stored in the culture collection of the Laboratory C-121, Biology Institute, UNAM, headed by Dr. Patricia Velez, and are fully available for research upon request.
Quality assessment and assembly of the ITS region and 16S Sanger sequences from fungal and bacterial isolates was performed using the finishing tool Consed version 29.0 (Ewing & Green, 1998; Ewing et al., 1998; Gordon et al., 2001). For the taxonomic assignment, sequence homology was evaluated through the comparison against the UNITE database for fungi (Kõljalg et al., 2020; Nilsson et al., 2019), and sequences from type material of the National Center of Biotechnology Information GenBank database using the BLAST algorithm for bacteria through a BLAST search (Abarenkov et al., 2010; Kõljalg et al., 2013). Sequence similarity for defining OTUs was set with a cut-off value of 98-100% for presumed species, 94-97% for genus level and 80-93% for order level. For conflicting hits, the lowest common rank level was used (Peršoh et al., 2010; Table 1). The sequences were deposited in GenBank under the accession numbers MT108978-MT109012 for fungi (Table 1), and MZ048754-MZ048770 for bacteria (Table 2).
Statistical analyses
We evaluated the relationship between microbial community structure and the following biogeochemical data retrieved from Velez et al. (2021; synchronously collected from the exact same sampling sites): 1) soil physicochemical properties: pH, NH4+, TC, total nitrogen and phosphorus (TN and TP respectively), DOC, dissolved nitrogen and phosphorus (DON and DOP respectively) and forms of carbon, nitrogen, and phosphorus contained in microbial biomass (Cmic, Nmic and Pmic respectively), and 2) 6 soil exoenzymes: β-1,4-glucosidase (BG), cellobiohydrolase (CBH), β-1,4-N-acetylglucosaminidase (NAG), phosphomonoesterase (AP), phosphodiesterase (APD), and POX. The raw data matrix was normalized using Z scores. We evaluated clustering patterns among sampling sites based on the culturable microbial community and environmental variables with the “hclust” function in ade4 v1.7-13 package in R (Dray & Dufour, 2007). A Principal Component Analysis (PCA) and a Spearman correlation test between soil biogeochemical variables and enzyme activities were conducted to select the variables for the subsequent multivariate analysis aiming to elucidate the relationships between biological assemblages of species and their environment. For the PCA, we considered as informative the components that represented at least 85% of the accumulative variance; whereas for the Spearman correlation matrix, we defined an uncorrelated model by using a threshold of 0.85 (Booth et al., 1994). These analyses were computed in R software 3.6.0 (R Core Team, 2018) using FactoMineR version 2.1 (Lê et al., 2008). Next, a Canonical Correspondence Analysis (CCA) with the selected biogeochemical variables (pH, DOC, DON, DOP, NH4+, POX, NAG and AP) and species data was calculated using the R package vegan (Oksanen et al., 2009).

Results
Overall 101 axenic fungal isolates were obtained from the 9 soil subsamples, clustering into 35 OTUs. The OTUs belonged to the phyla Mortierellomycota (Mortierellaceae sp. and Mortierella turficola), and Ascomycota (33 OTUs). The Ascomycota represented the most abundant and diverse phylum in our samples; affiliated with 8 orders: Capnodiales (1 OTU), Diaporthales (3 OTUs), Dothideales (1 OTU), Eurotiales (3 OTUs), Glomerellales (1 OTU), Hypocreales (14 OTUs), Magnaporthales (1 OTU) and Pleosporales (6 OTUs). At the genus level, isolates of the Ascomycota belonged to 21 genera: Aspergillus, Aureobasidium, Beauveria, Cladosporium, Clonostachys, Diaporthe, Furcasterigmium, Fusariella, Gaeumannomyces, Ilyonectria, Mariannaea, Metarhizium, Parapyrenochaeta, Parengyodontium, Penicillium, Phomopsis, Setophaeosphaeria, Talaromyces, Tolypocla-
dium, Trichoderma and Wojnowiciella (Table 1). Among these, the dominant component was Tolypocladium geodes.
We isolated 170 bacterial strains out of which representative isolates with distinctly unique morphologies were identified based on the homology of the 16S rRNA gene region towards reference sequences from the NCBI database. A total of 17 OTUs were delimited within the Actinobacteria (Arthrobacter and Microbacterium), Firmicutes (Bacillus) and Proteobacteria (Pseudomonas). The most abundant elements were Pseudomonas and Bacillus representatives.
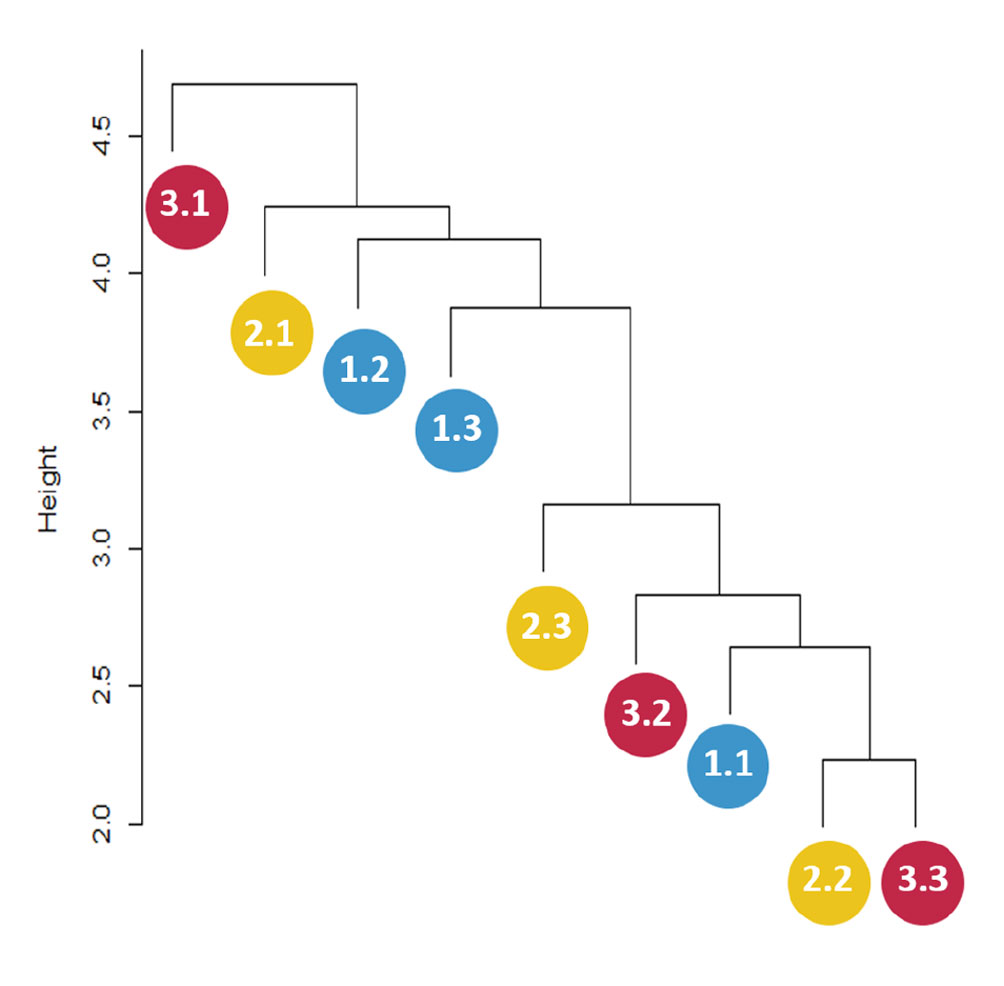
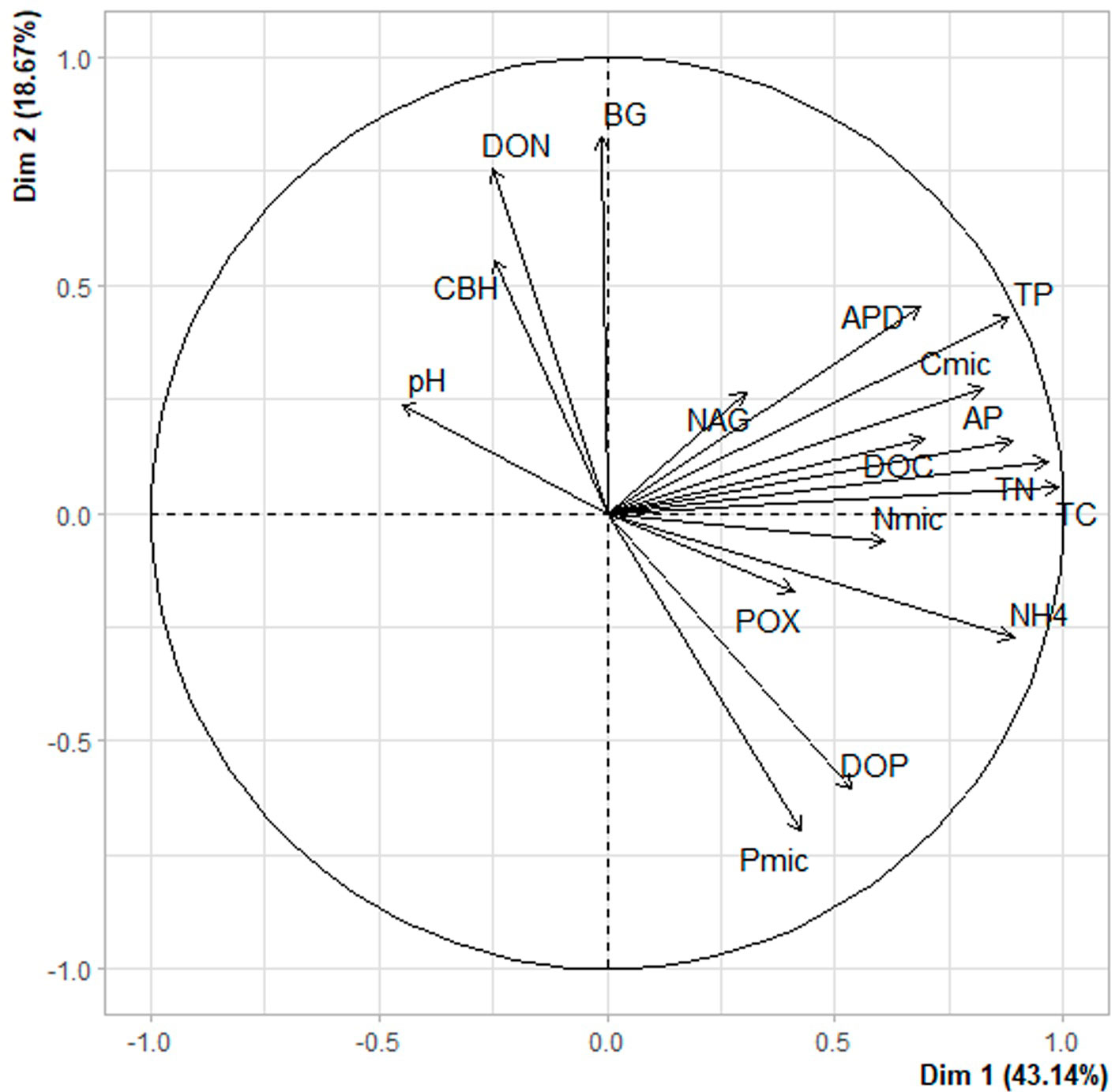
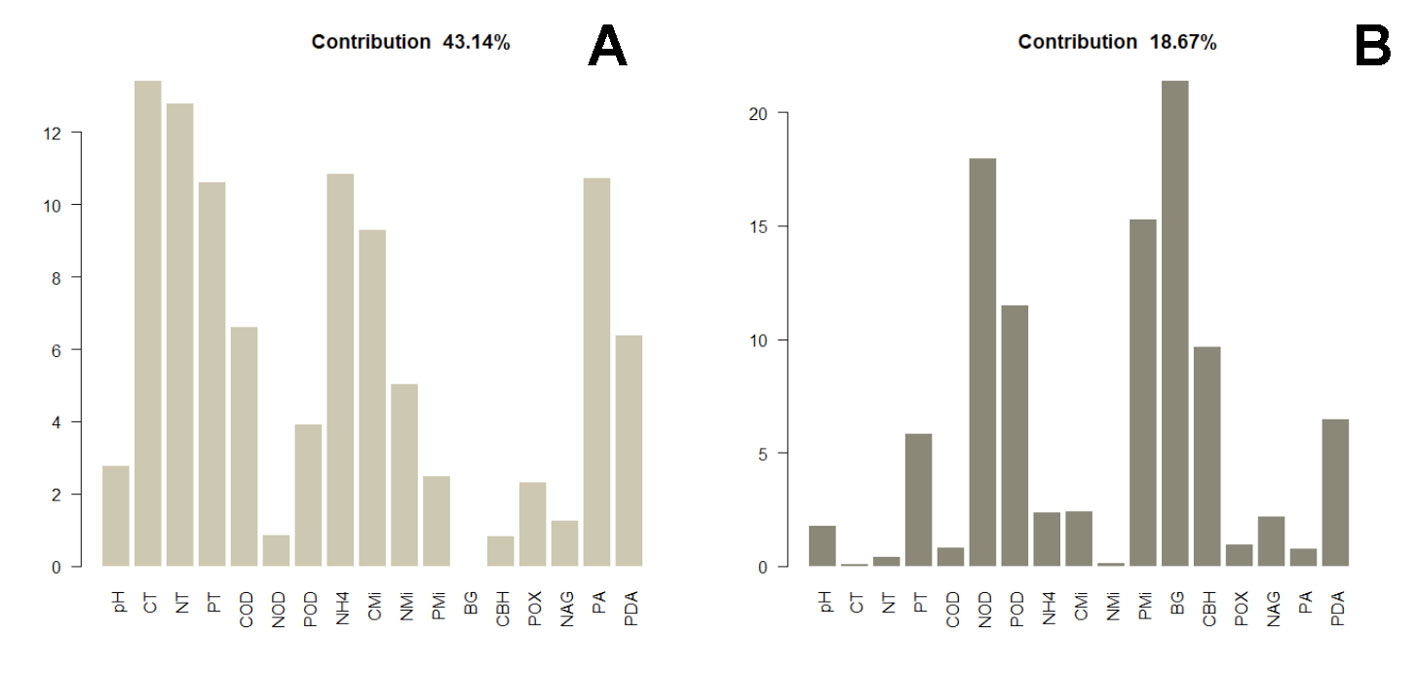
The distance dendrogram showed a sparse clustering among sampling sites (Fig. 3). Likewise, the PCA confirmed a considerable heterogeneity in the soil environmental data, with the first 2 ordination axes explaining 61.81% of the total variation (Fig. 4). The variables that most contributed to the first component were TC, TN, AP, NH4+ and TP; whereas for the second component BG, DON, Pmic and DOP showed the top contribution (Fig. 5). Spearman analysis showed significant correlations among biogeochemical variables, such as: NH4+, DOC and DOP, as well as AP, NAG, APD and POX (Supplementary material, Tables 1, 2).
Table 1
Fungal isolates obtained from soil samples collected in a pristine location of Mexican cloud forest. * Isolated from sick fronds of A. salvinii.
| Isolate | ITS1-5.8S-ITS2 | ||||
| OTU | Reference accession numbers | % Identity | e-value | Accession NCBI | |
| N10 | Aspergillus inflatus | MH859900 | |||
| MH859521 | |||||
| MH859519 | |||||
| AJ608959 | 99 | 0 | MT108998 | ||
| AF033393 | |||||
| T8 | Aureobasidium pullulans | MT882127 | |||
| MH864403 | |||||
| JX188099 | 100 | 0 | MT109010 | ||
| EU272483 | |||||
| MF062189 | |||||
| M22_T13 | Beauveria sp. | AY532003 | |||
| HQ880820 | |||||
| MH865206 | 99 | 0 | MT108987 | ||
| MH862139 | |||||
| HQ880819 | |||||
| N3_T2_T4_T9_M6 | Cladosporium sp. | MN543985 | |||
| MN543962 | |||||
| MN543951 | 100 | 0 | MT109002 | ||
| MN521809 | |||||
| MN518420 | |||||
| M5_2A | Clavicipitaceae sp. | MN905773.1 | |||
| HM030580 | |||||
| MH864652.1 | 100 | 0 | MT108996 | ||
| MH859547.1 | |||||
| AB709835.1 | |||||
| M19_Tube 12 | Clonostachys rosea* | MN511326 | |||
| KX421414 | |||||
| HM052817 | 99 | 0 | MT109000 | ||
| KM265525 | |||||
| MH859090 | |||||
| T19_N16 | Diaporthaceae sp. | MH864503.1 | 99 | 0 | MT108997 |
| MH299960.1 | |||||
| MH299958.1 | |||||
| MH020798.1 | |||||
| FN597586.1 | |||||
| M20_M26 | Diaporthe sp. | MF435154 | |||
| MF435146 |
| Table 1. Continued | |||||
| Isolate | ITS1-5.8S-ITS2 | ||||
| OTU | Reference accession numbers | % Identity | e-value | Accession NCBI | |
| MF435133 | 100 | 0 | MT108986 | ||
| MF435132 | |||||
| MF435131 | |||||
| M10_N11_M28 | Didymellaceae sp. | MH861244 | |||
| MN077427 | |||||
| JF817335 | 99 | 0 | MT108978 | ||
| JN207257 | |||||
| MF435134 | |||||
| M23 | Dothidiomycetes sp. | MN421894 | |||
| MN421889 | |||||
| MN421869 | 99 | 0 | MT108981 | ||
| KX640595 | |||||
| KX640594 | |||||
| M13 | Furcasterigmium furcatum | MH859660 | |||
| MH856099 | |||||
| AJ608973 | 99 | 0 | MT108980 | ||
| JF311914 | |||||
| LR590130 | |||||
| M2B | Fusariella sp. | EU687056 | 96 | ||
| KF800481 | 94 | ||||
| FJ820737 | 94 | 0 | MT108986 | ||
| MH859784 | 94 | ||||
| MH860688 | 93 | ||||
| M25 | Gaeumannomyces californicus | NR_155135.1 | 98.149 | ||
| NR_155133.1 | 97.799 | ||||
| KX306490.1 | 98.149 | 0 | MT108990 | ||
| KX306480.1 | 97.799 | ||||
| KX306482.1 | 97.269 | ||||
| T14 | Ilyonectria sp. | KP761750 | 100 | 0 | MT109005 |
| MK164179 | |||||
| KF895008 | |||||
| MF101382 | |||||
| LC133803 | |||||
| Tube 8_Tube 9 | Mariannaea sp. | MH863675 | |||
| MH862153 | |||||
| KM231758 | 100 | 0 | MT109001 | ||
| KM231757 | |||||
| KF767354 | |||||
| N14 | Metarhizium anisopliae | MH864642 | |||
| MH483803 | |||||
| KY786031 | 99 | 0 | MT109000 | ||
| EU307915 | |||||
| AF137059 | |||||
| M34 | Metarhizium carneum | MK164228 | |||
| MK164227 | |||||
| HQ392598 | 98 | 0 | MT109012 | ||
| MK387968 | |||||
| EU553292 | |||||
| M9 | Mortierellaceae sp. | MH860437 | |||
| MH860436 | |||||
| MH860435 | 99 | 0 | MT108997 | ||
| MH860122 | |||||
| MH860121 | |||||
| M17 | Mortierella turficola | EF521229 | |||
| AM292200 | |||||
| EU240043 | 99 | 0 | MT108982 | ||
| JX976025 | |||||
| JX975952 | |||||
| T11 | Nectriaceae sp. | KP265346 | 97 | ||
| MT534189 | 96 | ||||
| HQ897787 | 96 | 0 | MT109004 | ||
| HQ897787 | 96 | ||||
| AM410602 | 96 | ||||
| N13 | Parapyrenochaeta acaciae | KX228265 | 100 | ||
| KF673765 | 99 | ||||
| MK441755 | 95 | 0 | MT108999 | ||
| KX147607 | 98 | ||||
| KX147606 | 98 | ||||
| M14 | Parengyodontium album | MK834516 | 99 | 0 | MT109008 |
| MW187752 | |||||
| MW077094 | |||||
| MT672589 | |||||
| MT626052 | |||||
| M1A_M1B | Parengyodontium album | MK719933 | |||
| MH860372 | |||||
| LC092885 | 100 | 0 | MT108985 | ||
| LC092884 | |||||
| LC092882 | |||||
| N30_T5 | Penicillium sp. | NR_077153 | |||
| MN515068 | |||||
| MN511336 | 100 | 0 | MT109003 | ||
| MN371392 | |||||
| MT872087 | |||||
| M18_M3 | Phomopsis sp. | MF185326 | |||
| EU002915 | |||||
| MF185359 | 99 | 0 | MT108983 | ||
| MF185341 | |||||
| MF185334 | |||||
| T22 | Pleosporales sp. 1 | FM178244 | 99 | ||
| FM178246 | 98 | ||||
| MK066907 | 99 | 0 | MT109008 | ||
| MH931265 | 99 | ||||
| MH844084 | 99 | ||||
| M24 | Pleosporales sp. 2 | MH935005 | 100 | ||
| KY367514 | 99 | ||||
| KT309810 | 100 | 0 | MT108989 | ||
| MH861839 | 99 | ||||
| KY940787 | 99 | ||||
| Tube 6 | Pleosporomycetidae sp. | KJ591760 | 96 | ||
| KY454761 | 96 | ||||
| LT623218 | 96 | 0 | MT108992 | ||
| KF811432 | 95 | ||||
| JQ388267 | 95 | ||||
| M11 | Setophaeosphaeria hemerocallidis | KJ869161 | 99 | ||
| KX515692 | 97 | ||||
| KX515688 | 97 | 0 | MT108979 | ||
| KX515679 | 97 | ||||
| KX515674 | 97 | ||||
| M28 | Talaromyces wortmannii | MK020174 | 100 | 0 | MT108991 |
| KF984826 | |||||
| KF984825 | |||||
| KF984824 | |||||
| KF984823 | |||||
| M35_M27_M30_M33_T20_M15_M32 | Tolypocladium geodes | MH859919 | |||
| KU556539 | 99 | 0 | MT108995 | ||
| JX507694 | |||||
| T16B_T17 | Trichoderma sp. 1 | MN516473 | |||
| MN516472 | |||||
| MN186861 | 100 | 0 | MT109006 | ||
| MN186859 | |||||
| MK871069 | |||||
| Tube 4 | Trichoderma sp. 2 | MN518401 | |||
| MN516457 | |||||
| MN516456 | 100 | 0 | MT109011 | ||
| MN516454 | |||||
| MN516452 | |||||
| T6 | Trichoderma koningii | Z79628 | |||
| X93983 | |||||
| MN516479 | 100 | 0 | MT109010 | ||
| MN516476 | |||||
| MN516475 | |||||
| M29 | Wojnowiciella dactylidis | LT990661 | |||
| LT990659 | |||||
| LT990658 | 99 | 0 | MT108993 | ||
| MK442631 | |||||
| KF800363 |
Table 2
Bacterial isolates obtained from soil samples collected in a pristine location of Mexican cloud forest.
| Isolate | 16S | ||||
| OTU | Accession NCBI | % Identity | e-value | Accession
NCBI |
|
| 28.P | Arthrobacter sp. | MW227493.1 | 100 | ||
| NR_133969.1 | 98.45 | 0 | MZ048754 | ||
| JX949648.2 | 98.01 | ||||
| MN080869.1 | 98.01 | ||||
| NR_170399.1 | 98.01 | ||||
| C4 | Bacillus sp. | CP009692.1 | 100 | ||
| NR_113990.1 | 100 |
| Table 2. Continued | |||||
| Isolate | 16S | ||||
| OTU | Accession NCBI | % Identity | e-value | Accession NCBI | |
| AM747229.1 | 100 | 0 | MZ048770 | ||
| NR_115993.1 | 100 | ||||
| NR_036880.1 | 100 | ||||
| N10 | Microbacterium sp. | MK424288.1 | 100 | ||
| NR_042263.1 | 100 | ||||
| MT760166.1 | 99.47 | 0 | MZ048756 | ||
| NR_117603.1 | 99.47 | ||||
| MT760185.1 | 97.87 | ||||
| 47.P | Planococcaceae sp. 1 | NR_113837.1 | 99.78 | ||
| NR_029233.1 | 99.78 | ||||
| X68415.1 | 99.78 | 0 | MZ048755 | ||
| NR_113752.1 | 99.57 | ||||
| MT760068.1 | 99.57 | ||||
| N12 | Planococcaceae sp. 2 | NR_025628.1 | 98.22 | 1.00E-165 | |
| NR_025627.1 | 98.22 | 1.00E-165 | MZ048757 | ||
| NR_109749.1 | 97.63 | 2.00E-162 | |||
| NR_041521.1 | 97.63 | 2.00E-162 | |||
| NR_118296.1 | 97.33 | 1.00E-160 | |||
| N13 | Planococcaceae sp. 3 | NR_025627.1 | 97.93 | 0 | |
| NR_025628.1 | 97.67 | 0 | MZ048758 | ||
| NR_025029.1 | 95.61 | 4.00E-175 | |||
| NR_041521.1 | 95.09 | 9.00E-172 | |||
| NR_144702.1 | 94.07 | 2.00E-164 | |||
| 46.P1 | Planococcaceae sp. 4 | NR_116601.1 | 97.86 | ||
| CP016539.2 | 97.69 | 0 | MZ048763 | ||
| CP013659.2 | 97.69 | ||||
| LC379145.1 | 97.69 | ||||
| NR_113814.1 | 97.69 | ||||
| 50.P2 | Planococcaceae sp. 5 | KU886574.1 | 94.93 | ||
| NR_171442.1 | 94.93 | 0 | MZ048764 | ||
| NR_134133.1 | 94.8 | ||||
| CP016534.2 | 94.74 | ||||
| CP016539.2 | 94.75 | ||||
| T2 | Planococcaceae sp. 6 | NR_113752.1 | 99.77 | ||
| MT760068.1 | 99.77 | 0 | MZ048769 | ||
| MT757992.1 | 99.77 | ||||
| NR_113837.1 | 99.08 | ||||
| NR_036942.1 | 99.08 | ||||
| T11 | Pseudomonas sp. 1 | LT629778.1 | 100 | ||
| CP062253.1 | 99.69 | 0 | MZ048759 | ||
| CP029608.1 | 99.54 | ||||
| KT321658.1 | 99.54 | ||||
| CP062252.1 | 99.54 | ||||
| 29.P1 | Pseudomonas sp. 2 | MT027239.1 | |||
| NR_103934.2 | 99.66 | 0 | MZ048760 | ||
| NR_148295.1 | |||||
| NR_134795.1 | |||||
| LK021121.2 | |||||
| N8 | Pseudomonas sp. 3 | NR_148295.1 | 99 | ||
| MW111151.1 | 98.67 | 0 | MZ048761 | ||
| LR134290.1 | 98.01 | ||||
| LC507444.1 | 97.84 | ||||
| NR_134795.1 | 97.84 | ||||
| 40.P | Pseudomonas sp. 4 | LT629790.1 | 99.32 | ||
| LC409077.1 | 99.01 | 0 | MZ048762 | ||
| MZ099645.1 | 99.01 | ||||
| LC409075.1 | 98.94 | ||||
| NR_025102.1 | 98.86 | ||||
| C1 | Pseudomonas sp. 5 | JX545210.1 | 99.51 | ||
| LC595308.1 | 99.51 | 0 | MZ048767 | ||
| MK680061.1 | 99.18 | ||||
| MG719526.1 | 99.18 | ||||
| CP009533.1 | 99.18 | ||||
| C2 | Pseudomonas sp. 6 | LC500864.1 | 100 | ||
| LC548100.1 | 99.86 | 0 | MZ048768 | ||
| KX186943.1 | 99.86 | ||||
| KX186942.1 | 99.86 | ||||
| KX186936.1 | 99.86 | ||||
| N251 | Xanthomonadaceae sp. | NR_121739.1 | 98.73 | ||
| CP007597.1 | 98.73 | 0 | MZ048765 | ||
| MW629800.1 | 98.73 | ||||
| KY020782.1 | 98.36 | ||||
| NR_028930.1 | 98.37 | ||||
| T17 | Xanthomonadaceae sp. | NR_121739.1 | 99.47 | ||
| CP007597.1 | 99.47 | 0 | MZ048766 | ||
| MW629800.1 | 99.47 | ||||
| NR_028930.1 | 99.47 | ||||
| AJ293463.1 | 99.47 |
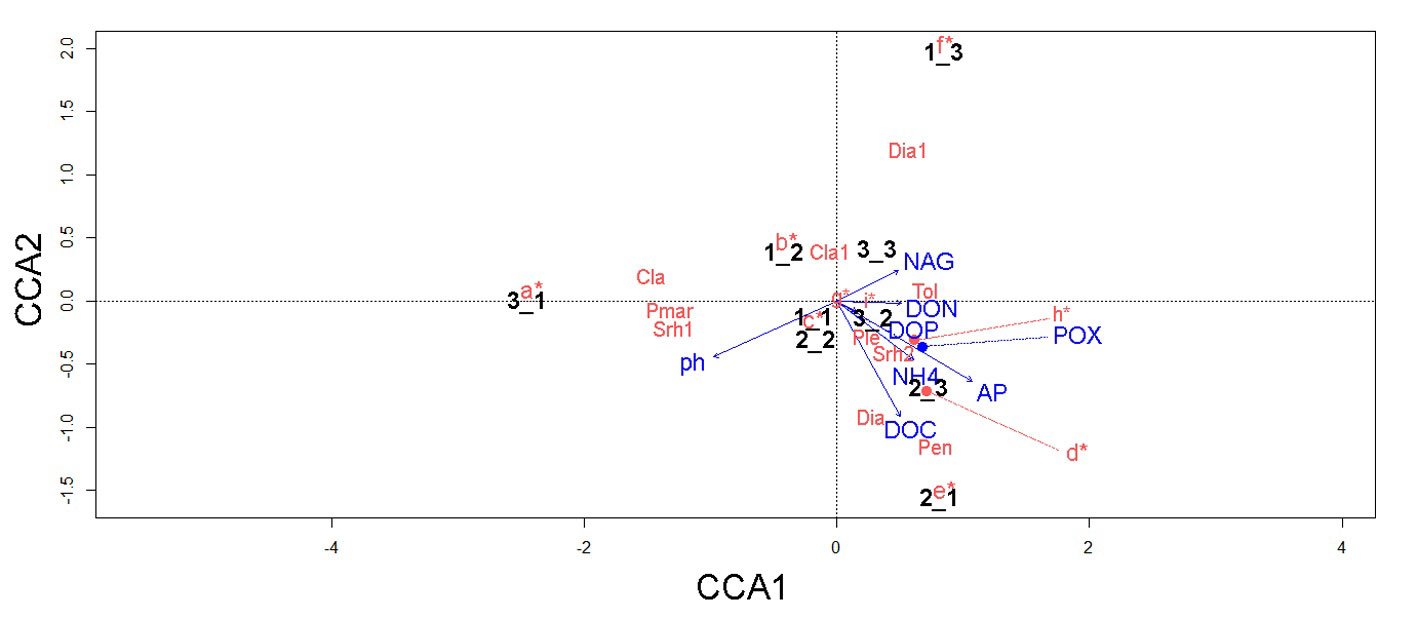
The CCA data suggested that the distribution of fungal and bacterial assemblages in the soil is strongly associated with key environmental variables (Fig. 6). For instance, we detected 5 relevant associations: 1) T. geodes and DON; 2) Penicillium sp., Diaporthaceae sp., and DOC; 3) Pleosporales sp. M24, Xanthomonadaceae sp. N251, and POX; and 4) M. turficola, Dothideomycetes sp., Metarhizium carneum, Wojnowiciella dactylidis, and NH4+ (linked to the samples collected near the fern A. salvinii).
Discussion
Soil microbiota, including bacteria and fungi, plays central roles in soil fertility and promotes plant health via complex cross-kingdom interactions. Nonetheless, microbial diversity in soils remains poorly understood at different spatial scales. Herein we report 52 microbial OTUs that represent several edaphic functional guilds at the small-scale. This culture-based approach provides the opportunity for a posteriori studies and the possibility of ex situ preservation of genetic resources in face of MCF imminent threats.
Compared with culture-dependent studies of soil fungal diversity in cloud forests at the large-scale (e.g., 90 samples across coffee plantations and MCF yielding to 415 species in Arias and Heredia-Abarca [2014]; and 20 samples from 4 forest fragments reporting 233 species in Arias and Heredia-Abarca [2020]), our results suggested moderate culturable diversity levels within a 10 × 10 × 10 m-transect. Remarkably, the occurrence fungal OTUs such as Trichoderma koningii and species of the genera Beauveria, Cladosporium, Penicillium and Trichoderma, agree with former reports on these taxa from conserved and fragmented cloud forest sites (Arias & Heredia-Abarca, 2014, 2020). In terms of prokaryotic diversity, the most abundant genera were Pseudomonas and Bacillus (both potentially phosphate solubilizer bacteria) in agreement with former work in the Santuario del Bosque de Niebla, a protected area of MCF in Veracruz State (Reverchon et al., 2019, 2020).
The obtained fungi included ubiquitous soil saprobes, as well as potential pathogens of insects, plants, and fungi. In this sense, the abundant isolation of entomopathogenic fungi such as T. geodes, Beauveria, Metarhizium, and Trichoderma members, agrees with previous reports from remnants of the original cloud forest in Mexico (Arias & Heredia-Abarca, 2014; Zarza et al., 2022) and may indicate strong antagonistic processes in soil communities (Zimmermann, 1993). In addition, these taxa may be implicated as an important component of edaphic nitrogen dynamics, by mobilizing nitrogen from hosts (e.g., insects) to the soil, resulting in increased nitrogen availability (Behie et al., 2012), in accordance to the observed high values of β-1,4-N-acetylglucosaminidase —chitinolytic enzyme involved in C and N-acquiring microbial activities that is highly correlated with fungal biomass (Miller et al., 1998; Parham & Deng, 2000; Sinsabaugh & Findlay, 1995).
Given the importance of prompt disease detection and identification of ethological agents in phytopathology, particularly for endemic species inhabiting fragile ecosystems (such as A. salvinii), as a marginal result we present the first report of Clonostachys rosea as a possible phytopathogen of A. salvinii. This fungus has been identified as a phytopathogen of numerous hosts including faba bean (Afshari & Hemmati, 2017), Gastrodia elata (Lee et al., 2020), soybean (Bienapfl et al., 2012) and the fern Sphaeropteris lepifera (Guu et al., 2010); remarkably causing disease under environmental conditions similar to MCF. In this work, we detected the occurrence of C. rosea in soil samples and on sick fronds of A. salvinii, alerting about a possible emerging disease that should be further monitored.
Overall, we did not detect sharp spatial patterns at the small-scale in the analyzed microbial communities, resembling previous observations on the marked spatial heterogeneity at this scale in soil communities (e.g., Nielsen et al., 2010). Particularly, the CCA results for bacteria depicted no particular influence of the tested biogeochemical factors on bacteria. This highlights the need of understanding how small-scale environmental heterogeneity underlies microbial species richness in MCF. Nonetheless, in accordance with our hypothesis, we observed that some microbial players were strongly associated with particular soil biogeochemical variables. For example, T. geodes (entomopathogen) was associated with DON. This is relevant, as Tolypocladium members are known as key players in denitrification processes (Jirout, 2015). Furthermore, the association between M. turficola —plant growth promoting fungus (Ozimek & Hanaka, 2021)—, M. carneum (entomopathogen), and W. dactylidis —potentially phytopathogenic (Marin-Felix et al., 2019)—, with NH4+ in samples collected near A. salvinii might indicate their contribution to the regulation of edaphic inorganic N in the proximities of this fern.
The copious isolation of antagonic OTUs such as T. geodes, Metarhizium spp., and M. turficola —taxa secreting siderophores, a peptide with potential for biological control of fungi and bacteria (Ozimek & Hanaka, 2021)—, agrees with former metabarcoding data (Velez et al., 2021), and suggests imperative in situ biotic regulatory processes of ecosystem functioning, which should be further confirmed by experimental work. In this sense, research on microbial interactions should shed light into building models to predict the outcome of community alterations and the effects of perturbations (Faust & Raes, 2012).
Soil microbial community in the examined pristine Mexican MCF was dominated by potentially entomopathogenic fungal taxa such as T. geodes, and theoretically phosphate solubilizer bacteria such as Pseudomonas and Bacillus spp. In accordance with our hypothesis, microbial assemblages were associated with soil biogeochemical variables such as DON, DOC, POX and NH4+. The lack of small-scale community structure patterns coupled to a strong environmental heterogeneity, even at the small-spatial scale, is of vital significance and should be considered for the development and application of in situ conservation strategies. This is the first report of C. rosea as a phytopathogen of A. salvinii, which could pose a threat to the communities of this emblematic plant species. In the future, the possible utilization of the herein isolated native microbial genetic resources ought to be probed to evaluate their response to shifting environmental conditions, as well as for their relationship with plant hosts.
Acknowledgements
This research work was financially supported by DGAPA-PAPIIT-UNAM IA201319 and IA206219. We thank Jaime Gasca-Pineda and Gabriel Merino for help provided during sample collection; Lidia I. Cabrera Martínez for technical support during molecular work at the Laboratorio de Sistemática Molecular del Departamento de Botánica (Instituto de Biología, UNAM); Laura Márquez and Nelly López for their assistance during sequencing procedures at the Laboratorio Nacional de Biodiversidad (Instituto de Biología, UNAM). We also acknowledge the authorities of the municipality of Santiago Comaltepec, Oaxaca, for the facilities to carry out fieldwork.
References
Abarenkov, K., Henrik, R., Larsson, K., Alexander, I. J., Eberhardt, U., Erland, S. et al. (2010). The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytologist, 186, 281–285. https://doi.org/10.1111/j.1469-8137.2009.03160.x
Afshari, N., & Hemmati, R. (2017). First report of the occurrence and pathogenicity of Clonostachys rosea on faba bean. Australasian Plant Pathology, 46, 231–234. https://doi.org/10.1007/s13313-017-0482-3
Alfaro-Sánchez, G. (2004). Suelos. In A. J. García-Mendoza, M. J. Ordóñez, & M. Briones-Salas (Eds.), Biodiversidad de Oaxaca (pp. 55–65). México D.F.: Instituto de Biología, UNAM/ Fondo Oaxaqueño para la Conservación de la Naturaleza/ World Wildlife Fund.
Alfonso-Corrado, C., Naranjo-Luna, F., Clark-Tapia, R., Campos, J. E., Rojas-Soto, O. R., Luna-Krauletz, M. D. et al. (2017). Effects of environmental changes on the occurrence of Oreomunnea mexicana (Juglandaceae) in a biodiversity hotspot cloud forest. Forests, 8, 1–15. https://doi.org/10.3390/f8080261
Arias, R. M., & Heredia-Abarca, G. (2014). Fungal diversity in coffee plantation systems and in a tropical montane cloud forest in Veracruz, Mexico. Agroforestry Systems, 88, 921–933. https://doi.org/10.33885/sf.2020.50.1290
Arias, R. M., & Heredia-Abarca, G. (2020). Diversity of soil culturable fungi in the tropical montane cloud forest of Veracruz, Mexico. Scientia Fungorum, 50, e1290. https://doi.org/10.33885/sf.2020.50.1290
Bąk, T. (2014). Triangular method of spatial sampling. Statistics in Transition, 15, 9–22.
Bazzaz, F. A. (1998). Tropical forests in a future climate: changes in biological diversity and impact on the global carbon cycle. In A. Markham (Ed.), Potential impacts of climate change on tropical forest ecosystems (pp. 177–196). Dordrecht: Springer. https://doi.org/10.1007/978-94-017-2730-3_7
Behie, S. W., Zelisko, P. M., & Bidochka, M. J. (2012). Endophytic insect-parasitic fungi translocate nitrogen directly from insects to plants. Science, 336, 1576–1577. https://doi.org/10.1126/science.1222289
Bienapfl, J. C., Floyd, C. M., Percich, J. A., & Malvick, D. K. (2012). First report of Clonostachys rosea causing root rot of soybean in the United States. Plant Disease, 96, 1700–1700. https://doi.org/10.1094/PDIS-06-12-0550-PDN
Booth, G. D., Niccolucci, M. J., & Schuster, E. G. (1994). Identifying proxy sets in multiple linear regression: an aid to better coefficient interpretation. Ogden, UT: U.S. Department of Agriculture, Forest Service, Intermountain Research Station.
Brown, N. A., Bass, C., Baldwin, T. K., Chen, H., Massot, F., Carion, P. W. C. et al. (2011). Characterisation of the Fusarium graminearum -wheat floral interaction. Journal of Pathogens, 2011, 626345. https://doi.org/10.4061/2011/626345
Bruijnzeel, L. A., Kappelle, M., Mulligan, M., & Scatena, F. N. (2010). Tropical montane cloud forests: state of knowledge and sustainability perspectives in a changing world. In L. A. Bruijnzeel, F. N. Scatena, & L. S. Hamilton (Eds.), Tropical montane cloud forest: science for conservation and management (pp. 691–740). New York: Cambridge University Press.
Bruijnzeel, L. A., Mulligan, M., & Scatena, F. N. (2011). Hydrometeorology of tropical montane cloud forests: emerging patterns. Hydrological Processes, 25, 465–498. https://doi.org/10.1002/hyp.7974
Bubb, P., May, I., Miles, L., & Sayer, J. (2004). Cloud forest agenda. Cambridge, UK: UNEP-WCMC.
Corrales, A., Xu, H., Garibay-Orijel, R., Alfonso-Corrado, C., Williams-Linera, G., Chu, C. et al. (2021). Fungal communities associated with roots of two closely related Juglandaceae species with a disjunct distribution in the tropics. Fungal Ecology, 50, 101023. https://doi.org/10.1016/j.funeco.2020.101023
de Boer, W. D., Folman, L. B., Summerbell, R. C., & Boddy L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiology Reviews, 29, 795–811. https://doi.org/10.1016/j.femsre.2004.11.005
De Long, J. R., Jackson, B. G., Wilkinson, A., Pritchard, W. J., Oakley, S., Mason, K. E. et al. (2019). Relationships between plant traits, soil properties and carbon fluxes differ between monocultures and mixed communities in temperate grassland. Journal of Ecology, 107, 1704–1719. https://10.1111/1365-2745.13160
del Mar Delgado-Serrano, M., Escalante, R., & Basurto, S. (2015). Is the community-based management of natural resources inherently linked to resilience? An analysis of the Santiago Comaltepec community (Mexico). Ager. Revista de Estudios sobre Despoblación y Desarrollo Rural, 18, 91–114.
Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 19, 11–15.
Dray, S., & Dufour, A. B. (2007). The ade4 package: Implementing the duality diagram for ecologists. Journal of Statistical Software, 22, 1–20. https://doi.org/10.18637/jss.v022.i04
Ewing, B., & Green, P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research, 8, 186–194. https://doi.org/10.1101/gr.8.3.186
Ewing, B., Hillier, L., Wendl, M. C., & Green, P. (1998). Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Research, 8, 175–185. https://doi.org/10.1101/gr.8.3.175
Faust, K., & Raes, J. (2012). Microbial interactions: from networks to models. Nature Reviews Microbiology, 10, 538–550. https://doi.org/10.1038/nrmicro2832
Foster, P. (2001). The potential negative impacts of global climate change on tropical montane cloud forests. Earth-Science Reviews, 55, 73–106. https://doi.org/10.1016/s0012-8252(01)00056-3
Gordon, D., Desmarais, C., & Green, P. (2001). Automated Finishing with Autofinish. Genome Research, 11, 614–625. https://doi.org/10.1101/gr.171401
Guu, J. R., Ju, Y. M., & Hsieh, H. J. (2010). Bionectriaceous fungi collected from forest in Taiwan. Botanical Studies, 51, 61–74.
Hamilton, L. S. (2009). Los bosques y el agua. Estudio temático elaborado en el ámbito de la evaluación de los recursos forestales mundiales 2005. Italia, Roma: FAO.
Heredia-Abarca, G., Arias, R. M., & Gómez, S. (2011). Hongos microscópicos: especies en restos vegetales y del suelo. In A. Cruz-Angón (Ed.), La biodiversidad en Veracruz estudio de estado. Volumen II (pp. 41–49). México D.F.: Comisión Nacional para el Conocimiento y Uso de la Biodiversidad/ Universidad Veracruzana/ Instituto de Ecología, A.C.
Horwath, W. R. (2017). The role of the soil microbial biomass in cycling nutrients. In K. R. Tate (Ed.), Microbial biomass. A paradigm shift in terrestrial biogeochemistry (pp. 41–66). London: World Scientific.
Idriss, E. E., Makarewicz, O., Farouk, A., Rosner, K., Greiner, R., Bochow, H. et al. (2002). Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology, 148, 2097–2109. https://doi.org/10.1099/00221287-148-7-2097
INEGI, Instituto Nacional de Estadística y Geografía. (2010). Compendio de información geográfica municipal. Santiago Comaltepec, Oaxaca. México D.F.: Instituto Nacional de Estadística y Geografía.
Jirout, J. (2015). Nitrous oxide productivity of soil fungi along a gradient of cattle impact. Fungal Ecology, 17, 155–163. https://doi.org/10.1016/j.funeco.2015.07.003
Karger, D. N., Kessler, M., Lehnert, M., & Jetz, W. (2021). Limited protection and ongoing loss of tropical cloud forest biodiversity and ecosystems worldwide. Nature Ecology & Evolution, 5, 854–862. https://doi.org/10.1038/s41559-021-01450-y
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bahram, M. et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology, 22, 5271–5277. https://doi.org/10.1111/mec.12481
Kõljalg, U., Nilsson, H. R., Schigel, D., Tedersoo, L., Larsson, K. H., May, T. W. et al. (2020). The taxon hypothesis paradigm-on the unambiguous detection and communication of taxa. Microorganisms, 8, 1910. https://doi.org/10.3390/microorganisms8121910
Krasilnikov, P., Gutiérrez, M. C., Ahrens, R. J., Cruz, C. O., Sedov, S., & Solleiro, E. (2013). The soils of Mexico. Dordrecht: Springer.
Lane, D. J. (1991). 16S/23S rRNA Sequencing. In E. Stackebrandt, & M. Goodfellow (Eds.), Nucleic acid techniques in bacterial systematic (pp.115–175). New York: John Wiley and Sons.
Lê, S., Josse, J., & Husson, F. (2008). FactoMineR: an R Package for Multivariate Analysis. Journal of Statistical Software, 25, 1–18. https://doi.org/10.18637/jss.v025.i01
Lee, S. A., Kang, M. J., Kim, T. D., & Park, E. J. (2020). First Report of Clonostachys rosea Causing Root Rot of Gastrodia elata in Korea. Plant Disease, 104, 3069–3069. https://doi.org/10.1094/PDIS-01-20-0148-PDN
Leija-Loredo, E. G., & Pavón, N. P. (2017). The northernmost tropical rain forest of the Americas: Endangered by agriculture expansion. Tropical Ecology, 58, 641–652.
Marin-Felix, Y., Hernández-Restrepo, M., Wingfield, M. J., Akulov, A., Carnegie, A. J., Cheewangkoon, R. et al. (2019). Genera of phytopathogenic fungi: GOPHY 2. Studies in Mycology, 92, 47–133. https://doi.org/10.1016/j.simyco.2018.04.002
Martínez, M. L., Pérez-Maqueo, O., Vázquez, G., Castillo-Campos, G., García-Franco, J., Mehltreter, K. et al. (2009). Effects of land use change on biodiversity and ecosystem services in tropical montane cloud forest of Mexico. Forest Ecology and Management, 258, 1856–1863. https://doi.org/10.1016/j.foreco.2009.02.023
McGuire, K. L., & Treseder, K. K. (2010). Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biology and Biochemistry, 42, 529–535. https://doi.org/10.1016/j.soilbio.2009.11.016
Miller, M., Palojärvi, A., Rangger, A., Reeslev, M., & Kjøller, A. (1998). The use of fluorogenic substrates to measure fungal presence and activity in soil. Applied and Environmental Microbiology, 64, 613–617. https://doi.org/10.1128/aem.64.2.613-617.1998
Mori, A. S., Isbell, F., & Seidl, R. (2018). β-Diversity, community assembly and ecosystem functioning. Trends in Ecology and Evolution, 33, 549–564. https://doi.org/10.1016/j.tree.2018.04.012
Nemergut, D. R., Schmidt, S. K., Fukami, T., O´ Neill, S. P, Bilinski, T. M., Stanish, L. F. et al. (2013). Patterns and processes of microbial community assembly. Microbiology and Molecular Biology Reviews, 77, 342–356. https://doi.org/10.1128/MMBR.00051-12
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J. et al. (2016). FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology, 20, 241–248. https://doi.org/10.1016/j.funeco.2015.06.006
Nielsen, U. N., Osler, G. H., Cambell, C. D., Neilson, R., Burslem, D. F., & Van der Wal, R. (2010). The enigma of soil animal species diversity revisited: the role of small-scale heterogeneity. Plos One, 5, e11567. https://doi.org/10.1371/journal.pone.0011567
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson, J., Jeppesen, T. S., Schigel, D. et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, 47, 259–264. https://doi.org/10.1093/nar/gky1022
Ochoa-Ochoa, L. M., Mejía-Domínguez, N. R., & Bezaury-Creel, J. (2017). Priorización para la conservación de los Bosques de Niebla en México. Ecosistemas, 26, 27–37. https://doi.org/10.7818/ECOS.2017.26-2.04
Oda, G. A. M., de Siqueira, M. F., Pires, A. D. S., & de Cássia Quitete-Portela, R. (2019). Micro- or macroscale? Which one best predicts the establishment of an endemic Atlantic Forest palm? Ecology and Evolution, 9, 7284–7290. https://doi.org/10.1002/ece3.5300
Oksanen, J., Kindt, R., Legendre, P., O’Hara, B., Simpson, G. L., Solymos, P. et al. (2009). vegan: Community Ecology Package, R package version 1.15-3. Retrieved from: http://cran-r-project.org/package=vegan
Ozimek, E., & Hanaka, A. (2021). Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture, 11, 7. https://doi.org/10.3390/agriculture11010007
Parham, J. A., & Deng, S. P. (2000). Detection, quantification and characterization of β-glucosaminidase activity in soil. Soil Biology and Biochemistry, 32, 1183–1190. https://doi.org/10.1016/S0038-0717(00)00034-1
Pascual-Mendoza, S., Clark-Tapia, R., Campos, J. E., Monsalvo-Reyes, A., Luna-Krauletz, M. D., Pacheco-Cruz, N. et al. (2020). Diversidad genética de Oreomunnea mexicana (Juglandaceae), relicta del bosque de niebla de Sierra Juárez, Oaxaca. México. Madera y Bosques, 26, e2621941. https://doi.org/10.21829/myb.2020.2621941
Peršoh, D., Melcher, M., Flessa, F., & Rambold, G. (2010). First fungal community analyses of endophytic ascomycetes associated with Viscum album ssp. austriacum and its host Pinus sylvestris. Fungal Biology, 114, 585–596. https://doi.org/10.1016/j.funbio.2010.04.009
Ponce-Reyes, R., Reynoso-Rosales, V. H., Watson, J. E. M., VanDer Wal, J., Fuller, R. A., Pressey, R. L. et al. (2012). Vulnerability of cloud forest reserves in Mexico to climate change. Nature Climate Change, 2, 448–452. https://doi.org/10.1038/nclimate1453
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for statistical computing, Vienna. https://www.R-project.org.
Reverchon, F., García, W., Guevara, E., Solís, I. A., Ferrera, O., & Lorea, F. (2019). Antifungal potential of Lauraceae rhizobacteria from a tropical montane cloud forest against Fusarium spp. Brazilian Journal of Microbiology, 50, 583–592. https://doi.org/10.1007/s42770-019-00094-2
Reverchon, F., Escudero-Osorio, Y. S., Morteo-Zavaleta, J., Guevara-Avendaño, E., & Ramírez-Vázquez, M. (2020). Inhibición de Fusarium solani por bacterias de la filósfera y rizósfera de árboles del bosque mesófilo de montaña. Biotecnología y Sustentabilidad, 5, 3–18. https://doi.org/10.57737/biotecnologiaysust.v5i1.738
Rinnan, R., & Bååth, E. (2009). Differential utilization of carbon substrates by bacteria and fungi in tundra soil. Applied and Environmental Microbiology, 75, 3611–3620. https://doi.org/10.1128/AEM.02865-08
Romaní, A. M., Fischer, H., Mille-Lindblom, C., & Tranvil, L. J. (2006). Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology, 87, 2559–2569. https://doi.org/10.1890/0012-9658(2006)87[2559:IOBAFO]2.0.CO;2
Rosas-Rangel, D. M., Mendoza, M. E., Gómez-Tagle, A., & Tobón-Marín, C. (2019). Advances and challenges in the knowledge on the tropical mountain cloud forests of Mexico. Madera Bosques, 25, e2511759. https://doi.org/10.21829/myb.2019.2511759
Rzedowski, J. (1996). Análisis preliminar de la flora vascular de los bosques mesófilos de montaña de México. Acta Botanica Mexicana, 35, 25–44. https://doi.org/10.21829/abm35.1996.955
Rzedowski, J., & Palacios-Chávez, R. (1977). The Mexican Engelhardtia (Oreomunnea) forest in the region of La Chinantla (Oaxaca, Mexico). A relic of the Cenozoic. Botanical Sciences, 36, 93–127. https://doi.org/10.17129/botsci.1161
Santillán, A., Cruz, S. Z., Calva, A., Ireta, A. D. R., & Bautista, J. (2020). Climatic water balance of mountain mesophilic forest in the huasteca. Ecosistemas y Recursos Agropecuarios, 7, e2016. https://doi.org/10.19136/era.a7n1.2016
Schulz, S., Brankatschk, R., Dümig, A., Kögel-Knabner, I., Schloter, M., & Zeyer, J. (2013). The role of microorganisms at different stages of ecosystem development for soil formation. Biogeosciences, 10, 3983–3996. https://doi.org/10.5194/bg-10-3983-2013
Sinsabaugh, R. L., & Findlay, S. (1995). Microbial production, enzyme activity, and carbon turnover in surface sediments of the Hudson River estuary. Microbial Ecology, 30, 127–141. https://doi.org/10.1007/BF00172569
Tapia-Torres, Y., & García-Oliva, F. (2013). La disponibilidad del fósforo es producto de la actividad bacteriana en el suelo en ecosistemas oligotróficos: una revisión crítica. Terra Latinoamericana, 31, 231–242.
Trejo, I. (2004). Clima. In A. J. García Mendoza, M. J. Ordónez, & M. Briones-Salas (Eds.), Biodiversidad de Oaxaca (pp. 67–85). México D.F.: Instituto de Biología, UNAM/ Fondo oaxaqueño para la conservación de la naturaleza/ World Wildlife Fund.
Velez, P., Tapia-Torres, Y., García-Oliva, F., & Gasca-Pineda, J. (2021). Small-scale variation in a pristine montane cloud forest: evidence on high soil fungal diversity and biogeochemical heterogeneity. PeerJ, 9, e11956. https://doi.org/10.7717/peerj.11956
Wagg, C., Dudenhöffer, J. H., Widmer, F., & Van Der Heijden, M. G. (2018). Linking diversity, synchrony and stability in soil microbial communities. Functional Ecology, 32, 1280–1292. https://doi.org/10.1111/1365-2435.13056
Warcup, J. H. (1960). Methods for isolation and estimation of activity of fungi in soil. In D. Parkinson, & J. Waid (Eds.), Ecology of soil Fungi (pp. 3–21). Liverpool: Liverpool University Press.
White, T. J., Bruns, T., Lee, S. J. W. T., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR protocols: a guide to methods and applications (pp. 315–322). New York: Academic Press.
Williams, G., Toledo, M., & Hernández, C. G. (2013). How heterogeneous are the cloud forest communities in the mountains of central Veracruz, Mexico? Plant Ecology, 214, 685–701. https://doi.org/10.1007/s11258-013-0199-5
Zak, D. R., Holmes, W. E., White, D. C., Peacock, A. D., & Tilman, D. (2003). Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology, 84, 2042–2050. https://doi.org/10.1890/02-0433
Zanne, A. E., Abarenkov, K., Afkhami, M. E., Aguilar-Trigueros, C. A., Bates, S., Bhatnagar, J. M. et al. (2020). Fungal functional ecology: bringing a trait-based approach to plant-associated fungi. Biological Reviews, 95, 409–433. https://doi.org/10.1111/brv.12570
Zarza, E., López-Pastrana, A., Damon, A., Guillén-Navarro, K., & García-Fajardo, L. V. (2022). Fungal diversity in shade-coffee plantations in Soconusco, Mexico. PeerJ, 10, e13610. https://doi.org/10.7717/peerj.13610
Zimmermann, G. (1993). The entomopathogenic fungus Metarhizium anisopliae and its potential as a biocontrol agent. Pesticide Science, 37, 375–379. https://doi.org/10.1002/ps.2780370410