Ricardo Pérez-Enríquez a, *, Alejandra Arciniega b, Noé Díaz-Viloria c, Salvador E. Lluch-Cota a
a Centro de Investigaciones Biológicas del Noroeste, S.C., Ave. Inst. Politécnico Nacional No. 195, 23096 La Paz, Baja California Sur, Mexico
b Universidad de La Rioja, Ave. de La Paz No. 93-10, Logroño, La Rioja, Spain
c Instituto Politécnico Nacional, Centro Interdisciplinario de Ciencias Marinas, Ave. Inst. Politécnico Nacional s/n, 23096 La Paz, Baja California Sur, Mexico
*Corresponding author: rperez@cibnor.mx (R. Pérez-Enríquez)
Received: 1 February 2022; accepted: 2 December 2022
Abstract
The study of planktonic mollusks is a relevant element to understand the dynamics of the benthic communities under present and future environmental conditions. We present the description of the biodiversity of planktonic gastropods at 2 sampling sites on the Pacific coast of the Baja California Peninsula, Mexico. Organisms collected from plankton tows at 2 locations (Cabo Tosco, n = 89 and La Bocana, n = 213) were sequenced for a portion of the 18S rDNA gene. High diversity was registered, with 71 Molecular Operational Taxonomic Units (MOTUs), which contrasts with the low phenotypic diversity of stereoscopic images. Differences in community composition between and within sampling sites indicate that planktonic gastropod distribution is not random but probably modulated by micro-environmental processes such as currents or biological events. The presence of non-gastropod sequences within some shells (n = 6) suggests their use as carriers of eggs or larvae of other taxa.
Keywords: Larval dispersal; Mollusca; Meroplankton; GenBank
© 2023 Universidad Nacional Autónoma de México, Instituto de Biología. This is an open access article under the CC BY-NC-ND license
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Diversidad de gasterópodos planctónicos del oeste de la Península de Baja California evaluada mediante secuencias de 18S ADNr
Resumen
El estudio de moluscos planctónicos es un elemento relevante para la comprensión de la dinámica de la comunidad bentónica bajo las condiciones ambientales actuales y futuras. Se presenta la descripción de la biodiversidad de gasterópodos planctónicos en 2 sitios de la costa del Pacífico de la península de Baja California, México. Organismos recolectados en arrastres planctónicos en 2 localidades (Cabo Tosco, n = 89 y La Bocana, n = 213) se secuenciaron para una fracción del gen 18S ADNr. Se registró una alta diversidad con 71 unidades taxonómicas moleculares operativas (MOTU), que contrasta con la baja diversidad fenotípica observada en imágenes de estereoscopio. Las diferencias en la composición de la comunidad entre y dentro de los sitios de muestreo, indican que la distribución de gasterópodos planctónicos no es aleatoria, sino que se encuentra, probablemente, modulada por procesos microambientales tales como corrientes o eventos biológicos. La presencia de secuencias de organismos que no son gasterópodos dentro de algunas conchas (n = 6) sugiere que son utilizadas por organismos de otros taxones como portadores de huevos o larvas.
Palabras clave: Dispersión larvaria; Mollusca; Meroplancton; GenBank
© 2023 Universidad Nacional Autónoma de México, Instituto de Biología. Este es un artículo Open Access bajo la licencia CC BY-NC-ND
Introduction
Planktonic dispersal in the marine environment is one of the leading connectivity processes among populations of meroplanktonic and holoplanktonic gastropods (Crocetta et al., 2020; Roegner, 2000), and is a relevant element of the trophic dynamics of ecosystems and indirect indicators of the health of adult populations (Campos & Landaeta, 2016; Chávez-Villegas et al., 2014). Planktonic dispersal study will aid in a better understanding of the molluskan community’s behavior under present and future environmental conditions (e.g., Aceves-Medina et al., 2020; Molina-González et al., 2018). For this purpose, baseline studies on the species composition that rely on the use of genetic markers are a starting point.
Planktonic gastropod identification is usually made using their morphology through microscope observation. The use of genetic markers (the Cytochrome Oxidase Subunit I of the mitochondrial DNA, COI) for this purpose is more recent. It has been used to study the genetic connectivity of the marine caenogastropod Bursa scrobilator (Crocetta et al., 2020). Due to its high conservation, the 18S ribosomal nuclear DNA (18S rDNA) gene has been proposed for biodiversity studies (Ranjithkumar et al., 2018).
Planktonic gastropods have been quite extensively studied worldwide (Bandel et al., 1997; Campos & Landaeta, 2016; Chávez-Villegas et al., 2014; Oliva-Rivera & Navarrete, 2007). However, in northwestern Mexico, the main efforts have been in the study of the species composition of holoplanktonic mollusks in the Gulf of California (Angulo-Campillo et al., 2011) and the western coast of the Baja California Peninsula (BCP) (Aceves-Medina et al., 2020; Molina-González et al., 2018; Moreno-Alcántara et al., 2020; Sánchez-Hidalgo y Anda, 1989), characterizing these regions as highly diverse. However, there is no available information about meroplanktonic gastropods in the region. Therefore, this work presents the first record of the planktonic gastropod biodiversity at 2 sites of the western coast of the BCP obtained by genetic markers.
Materials and methods
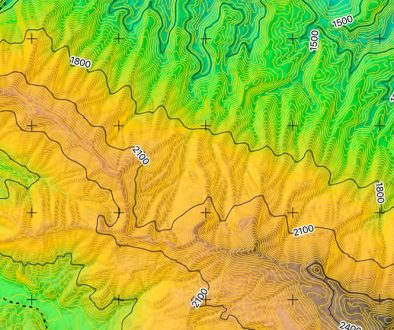
The sampling design originally intended to evaluate the presence of abalone larvae at 2 sites on the western coast of the Baja California Peninsula: La Bocana (LB: 26°45’1.9” N, 113°42’20.9” W) and Cabo Tosco (CT: 24°18’26.2” N, 111°42’36.6” W) (Fig. 1). However, we focused our study on other mollusks, as abalone larvae were not found. Sampling was done on December 1st, and 3rd, 2009 for LB and CT, respectively. At LB, the sampling area was located about 4.5 km southward, with an average depth of 10 m (Fig. 1). The rocky bottom floor is mostly flat with several small stone promontories composed of boulders, crevices, and a high abundance of macroalgae, mainly Eisenia arborea (Areschoug, 1876). CT is located at the southernmost part of Isla Margarita, and the sampling site was just 150 m off the coastline (Fig. 1), with a depth of approximately 3 m. The bottom is composed of large rocks with abundant crevices, surrounded by soft bottom but low macroalgae coverage. These sites were selected because they represent a high abalone productive area (LB) and the southern extreme of abalone species distribution (CT) (León-Carballo & Muciño-Díaz, 1996). The 8-day mean surface temperature in the first week of December 2009 at each site, which includes the sampling period and was obtained from the Aqua MODIS satellite —NPP, 0.0125°, West U.S., day time (11 microns), Simons (2022)— was 21 °C and 24 °C, respectively (Fig. 1).
At LB, there were 3 towing stations: 1) surface and 2) at 3-4 m depth at the same geographical position (LB-01 and LB-10, respectively), and 3) at the surface, approximately 1 km to the west (LB-07) of the previous position. At CT, there was one towing station (CT-01) at the surface, in a semi-protected area on the southwestern side of Isla Margarita. Tows followed a circular trajectory at a speed of 2.5 knots for 5 min. The plankton net was conical (0.45 m mouth diameter and 180 μm mesh size), with a calibrated flowmeter and a collection cup also with a 180 μm mesh size. Samples were transferred to a 500 mL plastic flask and preserved in 80% ethanol.
The biomass from the plankton tows was estimated by displaced volume (Beers, 1976) and standardized to mL of biovolume per 1,000 m3 of filtered seawater (Smith & Richardson, 1979). Due to high biomass at each towing station, the samples were fractionated to ¼ with a Folsom splitter. Using a stereoscope at 35×, following the description of Courtois de Viçose et al. (2007) for Haliotis tuberculata abalone larvae, gastropod mollusks with egg or globular shapes were separated (Bandel et al., 1997). A total of 384 individuals were separated, set in slides with wells (1 individual per well), and photo-documented (Leica Application Suite EZ, ver. 1.5.0). Individuals were transferred to 96-well PCR plates containing 70% ethanol.

DNA extraction was done following the protocol of Selvamani et al. (2001), with slight modifications. Before extraction, individuals were crushed with a plastic pestle and dried at ambient temperature. Samples were incubated in 18 μL of lysis buffer (Tris-HCl 10 mM, pH 8.3, KCl 50 mM, Tween-20 0.05%) and 2 μL de Proteinase-K (Sigma®, 2.5 mg/mL) at 55 °C for 24 h. After this time, the proteinase was inactivated by incubating the samples at 95 °C for 10 min. The plates were preserved at -20 °C until the PCR was performed.
A fragment of the 18S rDNA gene was amplified with primers 18SHal2F (5’-TTGGATAACTGTGGTAATTCTAGAGC) and 18SHal2R (5’-CCGGAATCGAACCCTGAT) (Aranceta-Garza et al., 2011). The reactions contained 0.2 mM de dNTPs (Invitrogen), 0.5 μM of each primer, PCR buffer 1× (Invitrogen), 1.5 mM MgCl2, 0.05 U Platinum® Taq DNA polymerase (Invitrogen), 2 μL of DNA and completed to 30 μL with MilliQ® water. PCR conditions were: 1 cycle at 94 °C for 4 min, 30 cycles at 94 °C for 45 s, 57 °C for 45 s, 72 °C for 45 s, and a final extension at 72 °C for 10 min in a thermal cycler (C1000, Bio-Rad). PCR products were verified in a 1.5% agarose gel stained with SYBR gold (Promega), visualized in a UV transilluminator (UVP BioDoc-it Imaging System).
PCR products of approximately 220 bp were individually sequenced either in forward and reverse (LB-07) or in forward only (LB-01, LB-10, CT-01) (Macrogen, Korea). The sequences were manually revised for quality and edited with ChromasPro v. 2.1.9 (Technelysium Pty Ltd). The edited sequences were analyzed to determine the number of similar genotypes with the software DnaSP v. 6.12.03 (Rozas et al., 2017). Identical sequences (100% match) were organized into Molecular Operational Taxonomical Units (MOTUs), for which the putative identity was obtained by a local alignment analysis with BLAST (Altschul et al., 1990). MOTUs were identified either to family, genus, or species level based on the hits with the highest similarity percentage (> 95%) and lowest E value (probability of the sequence match being random < 1 × 10-90). Nomenclature follows the NCBI (National Center for Biotechnology Information) Taxonomy database (Schoch, 2011).
The MOTUs sequences were analyzed as haplotypes (most sequences were homozygotes) to construct a minimum spanning network (Bandelt et al., 1999). The software PopArt (Leigh & Bryant, 2015) was used to visualize genealogical relationships among towing stations and obtain genetic diversity estimators (nucleotide diversity and number of segregating and parsimony-informative sites). The MOTUs’ diversity among towing stations was estimated by the Shannon-Weaver index using the natural logarithm with the vegan package (Oksanen et al., 2020) in R ver. 4.1.1 (R Core Team, 2013), using MOTUs as putatively non-identified species.
Results
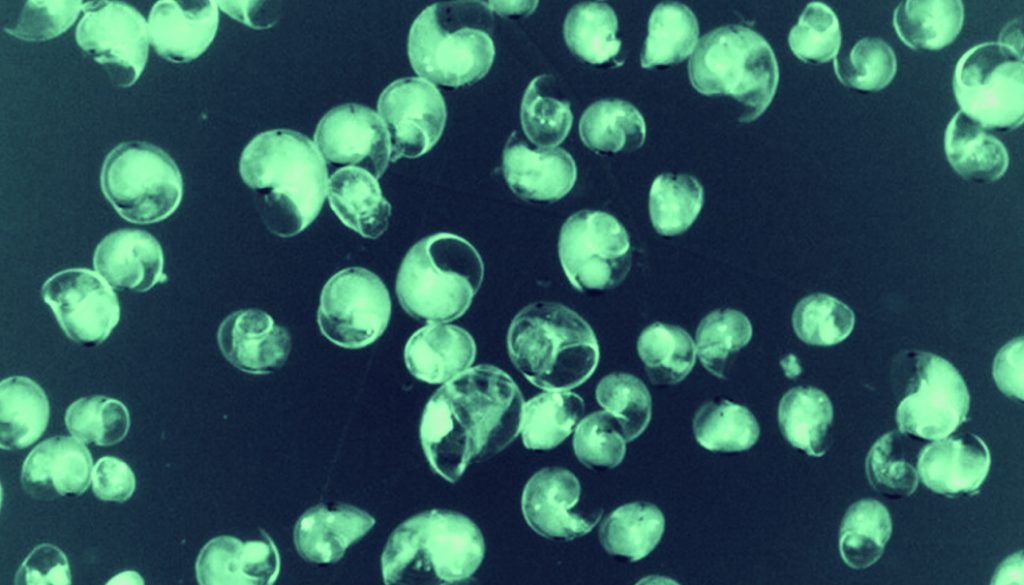
At LB, the plankton biomass at the mid-water tow was more than twice as high (LB-10, 1,222 mL 1,000 m–3) as at the 2 surface stations (LB-01 and LB-07, 491 and 557 mL 1,000 m–3, respectively). Surface biomass at CT (CT-01) was 883 mL 1,000 m–3. Planktonic gastropods, similar in shape, varied between 200 – 500 μm in width. As shells were translucent, it was possible to assess the presence of tissue within (Fig. 2).
Of a total of 384 individuals analyzed, 302 resulted in reliable DNA sequences matching gastropod mollusks (phylum Mollusca, class Gastropoda). The rest of the sequences were of low quality (n = 75) or matched other taxonomic groups (phylum Cnidaria, order Bivalvulida, and phylum Arthropoda, orders Euphausiacea and Calanoidea, n = 6) or showed no significant match to any species (n = 1). A total of 71 MOTUs were distinguished with matching identities higher than 95% and E-values lower than 1 × 10–100 in most sequences, putatively belonging to several families and orders of 5 subclasses (Tables 1, 2).
Table 1
Number of MOTUs and individuals per taxonomic group of the Class Gastropoda* at the 2 sampling sites.
| Subclass | Order | MOTU | Individuals |
| Caenogastropoda | Caenogastropoda incertae sedis | 2 | 3 |
| Littorinimorpha | 16 | 126 | |
| Neogastropoda | 5 | 8 | |
| Ptenoglossa (suborder) | 1 | 2 | |
| Heterobranchia | Aplysiida | 3 | 16 |
| Cephalaspidea | 6 | 11 | |
| Ellobiida | 1 | 2 | |
| Euthyneura (clade) | 1 | 1 | |
| Nudibranchia | 9 | 16 | |
| Pleurobranchida | 1 | 4 | |
| Pteropoda | 5 | 14 | |
| Heterostropha | 1 | 1 | |
| Sacoglossa | 9 | 34 | |
| Neritimorpha | Cycloneritida | 5 | 35 |
| Patellogastropoda | Patellogastropoda | 1 | 1 |
| Vetigastropoda | Lepetellida | 2 | 2 |
| Trochida | 2 | 5 | |
| Unclassified gastropod | 1 | 21 | |
| Total | 71 | 302 |
* Nomenclature from the Taxonomy browser of the NCBI database
(Schoch, 2011).
From the similarity analysis to GenBank sequences by BLAST, 23 MOTUs gave 100% identity. Identification to the species level was likely for 7 MOTUs (i.e., MOTUs 02, 05, 13, 33, 48, 61, and 62; Table 2). The remaining 16 are not certain as some of these MOTUs showed matching hits either to several genera (i.e., MOTUs 01, 03, 08, 10, 12, 15, 20, 22, 25, 28, 64, and 68; Table 2) or several species of a certain genus (i.e., MOTUs 29, 65, 66, and 67; Table 2).
Table 2
Individuals per sampling station, organized in Molecular Operational Taxonomic Units (MOTU) from the genetic identity analysis (BLAST) of the 18S rDNA gene sequences in the GenBank database. Genbank accession numbers are shown only for the MOTUs of highest identity percentage and lowest E-value hits. CT-01: Cabo Tosco; LB: La Bocana. BCP: Baja California Peninsula. Proporcionar tabla.
| MOTU | Sampling stations | Taxonomy of hits | GenBank hits characteristics | Notes on possible species in the study region (the Pacific coast of the BCP) based on the Global Biodiversity Information Facility web [https://www.gbif.org- Consulted: Jan, 14 2022] | ||||||||
| CT-01 | LB-01 | LB-10 | LB- 07 | Subclass | Order | Superfamily | Family | Genera and species (GenBank accession number) | E | % Identity | ||
| 01 | 2 | – | – | – | Caenogastropoda | Caenogastropoda incertae sedis | Abyssochry-soidea | Abyssochrysidae
Provannidae |
Abyssochrysos melanioides (AB930376)
Rubyspira osteovora (GQ290530) |
5×10-99 | 99.1 | There are no records of the genera Abyssochrysos or Rubyspira in the BCP |
| 02 | – | – | 1 | – | Epitonioidea | Epitoniidae | Alexania inawazai (AB930380) | 4×10-106 | 100 | There are no records of Alexania or other species of the genus in the BCP | ||
| 03 | 5 | – | – | – | Littorinimorpha | Littorinoidea Calyptraeoidea Vermetoidea | Lacunidae
Calyptraeidae Vermetidae |
Lacuna pallidula (AJ488686)
Crepidula fornicata (AY377660) Thylacodes adamsii (HQ833992) |
3×10-102 | 100 | The species Lacuna unifasciata (One-band lacuna) has been registered in the BCP. Several species of the genus Crepidula (American slipper limpet) have been registered in the BCP. Thylacodes adamsii (Scaly worm shell) has been registered in the north of the BCP | |
| 04 | 1 | 5 | 8 | 51 | Cypraeoidea | Cypraeidae | Erosaria erosa (KT753625)
Erronea erones (HQ833998) |
5×10-125 | 100 | Erosaria erosa and Erronea errones (Mistaken cowrie) are not registered in the BCP | ||
| 05 | – | 1 | – | – | Pterotra-cheoidea | Atlantidae | Atlanta sp. (MW203647) | 3×10-101 | 100 | Several species of the genus Atlanta have been registered in the Gulf of California (Angulo-Campillo et al., 2011), and the BCP (Aceves-Medina et al., 2020; Molina-González et al., 2018; Moreno-Alcántara et al., 2020). | ||
| 06 | – | – | – | 1 | Atlanta sp. (MW204226) | 2×10-123 | 99.6 | |||||
| 07 | – | 1 | – | – | Pterotra-cheoidea | Pterotracheidae | Firoloida desmarestia (MW204031) | 4×10-88 | 95.3 | The species Firoloida desmarestia have been registered in the Gulf of California (Angulo-Campillo et al., 2011), and the BCP ((Molina-González et al., 2018) | ||
| 08 | 2 | – | – | – | Rissooidea
Truncate-lloidea |
Rissoinidae
Hydrobiidae |
Phosinella clathrata (AB930392)
Rissoina fasciata (DQ916528) |
1×10-106 | 100 | No genera of families Rissoinidae or Hydrobiidae are registered in the BCP | ||
| 09 | – | 11 | 2 | – | Truncate-lloidea | Tateidae | Trochidrobia punicea (KT313223)
Novacaledonia numee (KT313210) Kanakyella gentilsiana (KT313209) and others |
5×10-99 | 99.1 | No genera of family Tateidae are registered in the BCP | ||
| 10 | – | 1 | – | – | Trochidrobia punicea (KT313223)
Novacaledonia numee (KT313210) Kanakyella gentilsiana (KT313209) and others |
9×10-102 | 100 | No genera of family Tateidae are registered in the BCP |
| Table 2. Continued | ||||||||||||
| MOTU | Sampling stations | Taxonomy of hits | GenBank hits characteristics | Notes on possible species in the study region (the Pacific coast of the BCP) based on the Global Biodiversity Information Facility web [https://www.gbif.org- Consulted: Jan, 14 2022] | ||||||||
| CT-01 | LB-01 | LB-10 | LB- 07 | Subclass | Order | Superfamily | Family | Genera and species (GenBank accession number) | E | % Identity | ||
| 16 | – | – | 1 | – | Caenogastropoda | Littorinimorpha | Phrantela daveyensis (KT313215) | 3×10-97 | 97.7 | No genus of Truncatelloidea is registered in the BCP | ||
| 11 | – | – | 1 | – | Tateidae
Hydrobiidae |
Phrantela daveyensis (KT313215)
Bythiospeum sp. (AF367664) |
1×10-93 | 97.7 | No genera of family Tateidae or Hydrobiidae are registered in the BCP | |||
| 12 | 1 | – | – | – | Caenogastropoda | Littorinimorpha | Vanikoroidea | Eulimidae | Pyramidelloides angustus (AB930386)
Hemiliostraca sp. (AB930383) |
6×10-104 | 100 | No species of Pyramidelloides or Hemiliostraca have been registered in the BCP |
| 13 | – | 8 | 6 | – | Balcis eburnea (AF120519) | 3×10-108 | 100 | The genus Balcis has been registered in the Gulf of California | ||||
| 14 | – | 1 | 6 | – | Melanella acicula (AB930381) | 1×10-101 | 98.2 | Several species of the genus Mellanela have been registered in the Gulf of California, and the BCP | ||||
| 15 | 5 | – | – | – | Hipponicidae | Cheilea pileopsis (AB930397)
Hipponicidae sp (MW204059) |
3×10-102 | 100 | The species Cheilea cepacea has been registered in the BCP | |||
| 18 | 7 | – | – | – | Vanikoridae | Vanikoro helicoidea (AB930395) | 2×10-100 | 97.7 | The species Vanikoro aperta has been registered in the Gulf of California but not in the BCP | |||
| 70 | – | 1 | – | – | Naticoidea | Naticidae | Sinum haliotoideum (FJ623466) | 8×10-99 | 98.1 | Several species of the genus Sinum have been registered in the BCP | ||
| 17 | – | – | – | 1 | Neogastropoda | Conoidea | Several families | Several species | 2×10-125 | 99.6 | No information available | |
| 19 | – | 1 | – | – | Buccinoidea | Several families | Several species | 4×10-100 | 99.5 | No information available | ||
| 20 | – | – | 2 | – | Several families | Several species | 5×10-105 | 100 | No information available | |||
| 21 | 2 | – | – | – | Muricoidea | Muricidae | Coralliophila caribaea (MW204229) | 2×10-103 | 99.5 | Several species of the genus Coralliophila have been registered in the Gulf of California, and the BCP | ||
| 22 | – | – | – | 2 | Caenogastropoda | Neogastropoda | Muricoidea | Muricidae | Several species | 6×10-124 | 100 | Several families of the genus Coralliophila have been registered in the Gulf of California, and the BCP |
| 23 | – | – | – | 2 | Ptenoglossa | Triphoroidea | Cerithiopsidae | Unclassified Cerithiopsidae | 3×10-122 | 99.6 | Several genera of family Cerithiopsidae are found in the BCP | |
| 24 | 7 | 3 | 1 | – | Akeroidea | Akeridae | Akera bullata (AY427502) | 3×10-113 | 99.6 | The species Akera maga has been registered in the BCP | ||
| 25 | 3 | – | – | – | Aplysiida | Aplysioidea | Aplysiidae | Stylocheilus longicauda (DQ093439)
Dolabrifera dolabrifera (DQ237960) |
3×10-112 | 100 | The species Stylocheilus ricketsii has been registered in the BCP and the Gulf of California. The species Dolabrifera dolabrifera has been registered in the Gulf of California | |
| 26 | – | 1 | 1 | – | Aplysia californica (XR_004859253) | 6×10-115 | 99.6 | The species Aplysia californica has been registered in the BCP | ||||
| 27 | 2 | – | 1 | – | Bulloidea | Bullidae | Bulla vernicosa (DQ923452) | 6×10-105 | 97.8 | Several species of the genus Bulla are registered in the BCP | ||
| 28 | – | – | 1 | – | Haminoeidae | Liloa mongii (MH933263)
Atys curta (DQ923459) |
9×10-113 | 100 | There are no records of Liloa or Atys for the BCP | |||
| 29 | – | – | 1 | – | Heterobranchia | Cephalaspidea | Diaphanoidea | Diaphanidae | Diaphana globosa (MH933321) | 1×10-107 | 98.2 | Diaphana californica has been registered in the BCP |
| 30 | 1 | – | – | – | Philinoidea | Cylichnidae | Acteocina lepta (MH933295) | 6×10-110 | 99.1 | Several species of the genus Acteocina have been registered in the BCP | ||
| 31 | 3 | – | – | – | Heterobranchia | Cephalaspidea | Philinoidea | Cylichnidae | Acteocina lepta (MH933295) | 7×10-90 | 99.5 | Several species of the genus Acteocina have been registered in the BCP |
| 32 | 1 | – | 1 | – | Laonidae | Laona confusa (MH933303) | 6×10-90 | 94.3 | Laona californica has been registered in a northern region of the BCP | |||
| 33 | – | 1 | 1 | – | Ellobiida | Ellobioidea | Ellobiidae | Melampus bullaoides (KM280980) | 7×10-114 | 100 | Several species of the genus Melampus have been registered in the BCP | |
| 35 | – | 1 | – | – | Euthyneura | Pyramide-lloidea | Pyramidellidae | Turbonilla elegantissima (GU331941) | 3×10-92 | 96.7 | Several species of the genus Turbonilla have been registered in the BCP | |
| 36 | – | 2 | – | – | Nudibranchia | Anadoridoidea | Polyceridae | Plocamopherus aurantinodulosa (EF534011) | 8×10-94 | 93.7 | There are no records of Plocamopherus for the BCP | |
| 37 | – | – | 1 | – | Fionoidea | Trinchesiidae | Phestilla sp. (MK088224) | 2×10-104 | 97.4 | Phestilla lugubris has been registered in the Gulf of California | ||
| 43 | – | – | 1 | – | Fionoidea | Eubranchidae | Eubranchus sanjuanensis (GQ326909) | 2×10-100 | 96.5 | Several species of the genus Eubranchus have been registered in the BCP | ||
| 38 | – | 1 | – | – | Dendrono-toidea | Dotidae | Doto columbiana (GQ326881) | 1×10-122 | 98.8 | Doto columbiana has been registered in a northern region of the BCP | ||
| 39 | – | – | 1 | – | Doto columbiana (GQ326881) | 3×10-118 | 97.6 | Doto columbiana have been registered in a northern region of the BCP | ||||
| 40 | – | 1 | – | – | Anadoridoidea | Onchidorididae | Corambe pacifica (KP340341) | 1×10-106 | 97.8 | Corambe pacifica has been registered in the BCP | ||
| 41 | – | – | 1 | – | Eudoridoidea | Chromodorididae | Mexichromis porterae (EF534014) | 3×10-108 | 97.8 | Two species of Mexichromis (M. antonii, M. tura) have been registered in the Gulf of California | ||
| 42 | – | – | 1 | 2 | Nudibranchia | Eudoridoidea | Chromodorididae | Diversidoris aurantinodulosa (EF534011) | 5×10-111 | 94.1 | There are no records of Diversidoris for the BCP | |
| 44 | 1 | – | 4 | – | Diversidoris aurantinodulosa (EF534011) | 4×10-92 | 94 | There are no records of Diversidoris for the BCP | ||||
| 45 | – | 2 | 2 | – | Pleurobranchida | Pleurobran-choidea | Pleurobranchidae | Berthella stellata (AY427495) | 9×10-73 | 90.5 | Berthella idiomorpha has been registered in the BCP | |
| 46 | – | 2 | – | – | Pteropoda | Thecosomata (Suborder) | Desmopteridae | Desmopterus papilio (GU969171) | 3×10-112 | 99.6 | Desmopterus papilio and D. pacificus have been registered in the Gulf of California (Angulo-Campillo et al., 2011) and the BCP (Molina-González et al., 2018; Sánchez-Hidalgo y Anda, 1994). | |
| 47 | – | 2 | – | 1 | Desmopterus papilio (GU969171) | 2×10-135 | 99.6 | |||||
| 48 | – | 3 | – | – | Desmopterus sp. (MW203406) | 2×10-115 | 100 | |||||
| 49 | – | 2 | 3 | – | Desmopterus sp. (MW203406) | 2×10-103 | 99.1 | |||||
| 50 | – | – | – | 1 | Desmopterus papilio (GU969171) | 9×10-134 | 99.3 | |||||
| 51 | – | 1 | – | – | Heterobranchia | Heterostropha | Pyramide-lloidea | Pyramidellidae | Hinemoa sp. (GU331936) | 2×10-105 | 97.8 | The genus Hinemoa has not been registered in the BCP |
| 52 | 1 | – | – | – | Limapontioidea | Caliphyllidae | Polybranchia sp. (MH375070) | 1×10-111 | 99.6 | Polybranchia mexicana has been registered in the BCP | ||
| 53 | – | 1 | – | – | Polybranchia sp. (MH375067) | 2×10-109 | 99.1 | Polybranchia mexicana has been registered in the BCP | ||||
| 54 | 1 | – | 1 | – | Polybranchia sp. (MH375070) | 4×10-96 | 96 | Polybranchia mexicana has been registered in the BCP | ||||
| 55 | – | 1 | – | – | Mourgona sp. (MH375064) | 1×10-96 | 96.4 | The genus Mourgona has not been registered in the BCP | ||||
| 56 | – | – | 1 | – | Mourgona sp. (MH375064) | 3×10-97 | 96.4 | The genus Mourgona has not been registered in the BCP | ||||
| 57 | 1 | 7 | 10 | 2 | Limapontiidae | Aplysiopsis minor (AB501328) | 1×10-127 | 98.9 | Aplysiopsis enteromorphae has been registered in the BCP | |||
| 58 | – | – | 1 | – | Aplysiopsis minor (AB501328) | 1×10-127 | 98.9 | Aplysiopsis enteromorphae has been registered in the BCP | ||||
| 34 | 5 | – | 1 | – | Heterobranchia | Sacoglossa | Stiliger smaragdinus (AB501324) | 3×10-97 | 96.4 | Stiliger fuscovittatus have been registered in the BCP | ||
| 59 | 1 | – | – | – | Oxynooidea | Oxynoeidae | Oxynoe viridis (AB501318)
Oxynoe antillarum (FJ917441) |
1×10-121 | 100 | Oxynoe aliciae, O. viridis and O. panamensis have been registered in the Gulf of California | ||
| 60 | 17 | – | – | – | Neritimorpha | Cycloneritida | Neritoidea | Neritidae | Nerita tessellata (FJ977654)
Nerita funiculata (DQ093429) |
7×10-104 | 99.1 | Nerita funiculata and Nerita tessellata have not been registered in the BCP |
| 61 | 13 | – | – | – | Nerita funiculata (DQ093429) | 4×10-106 | 100 | Nerita funiculata and Nerita tessellata have not been registered in the BCP | ||||
| 62 | 2 | – | – | – | Neritimorpha | Cycloneritida | Neritoidea | Neritidae | Nerita tessellata (FJ977654) | 4×10-106 | 100 | Nerita funiculata and Nerita tessellata have not been registered in the BCP |
| 63 | 1 | – | – | – | Neritidae | Nerita funiculata (DQ093429) | 5×10-105 | 99.5 | Nerita funiculata and Nerita tessellata have not been registered in the BCP | |||
| 64 | – | – | 2 | – | Phenacole-padidae | Phenacolepas osculans (AY923890) | 1×10-105 | 100 | Phenacolepas osculans has been registered in the Gulf of California | |||
| 65 | – | – | – | 1 | Patellogastropoda | Lottioidea | Lottiidae | Lottia gigantea (KP274858)
Lottia scabra (GQ160769) Lottia jamaicensis (FJ977633) |
6×10-151 | 100 | Several species of the genus Lottia have been registered in the BCP | |
| 66 | – | – | – | 1 | Vetigastropoda | Lepetellida | Fissurelloidea | Fissurellidae | Fissurella sp. (MK331687)
Fissurella virescens (MK322194) Fissurella volcano (HM775293) |
2×10-129 | 100 | Fissurella volcano and other Fissurella species have been registered in the BCP |
| 67 | – | 1 | – | – | Haliotoidea | Haliotidae | Haliotis corrugata (HM775288)
Haliotis fulgens (HM775289) |
3×10-102 | 100 | Haliotis corrugata and Haliotis fulgens have been registered in the BCP | ||
| 68 | 4 | – | – | – | Trochida | Trochoidea | Turbinidae | Megastrea undosa (KY766259)
Turbo sp. (KY766259) Lunella sp. (EU530105) |
1×10-95 | 100 | Several genera of family Turbinidae are found in the BCP | |
| 69 | – | 1 | – | – | Vetigastropoda | Trochida | Trochoidea | Phasianellidae | Tricolia pullus (AM048661) | 6×10-85 | 94.1 | There are no species of the genus Tricolia registered in the BCP |
| 71 | – | 15 | 6 | – | Gastropoda unclassified (MW203299) | 3×10-101 | 99.1 | |||||
| Σ | 89 | 78 | 70 | 65 |
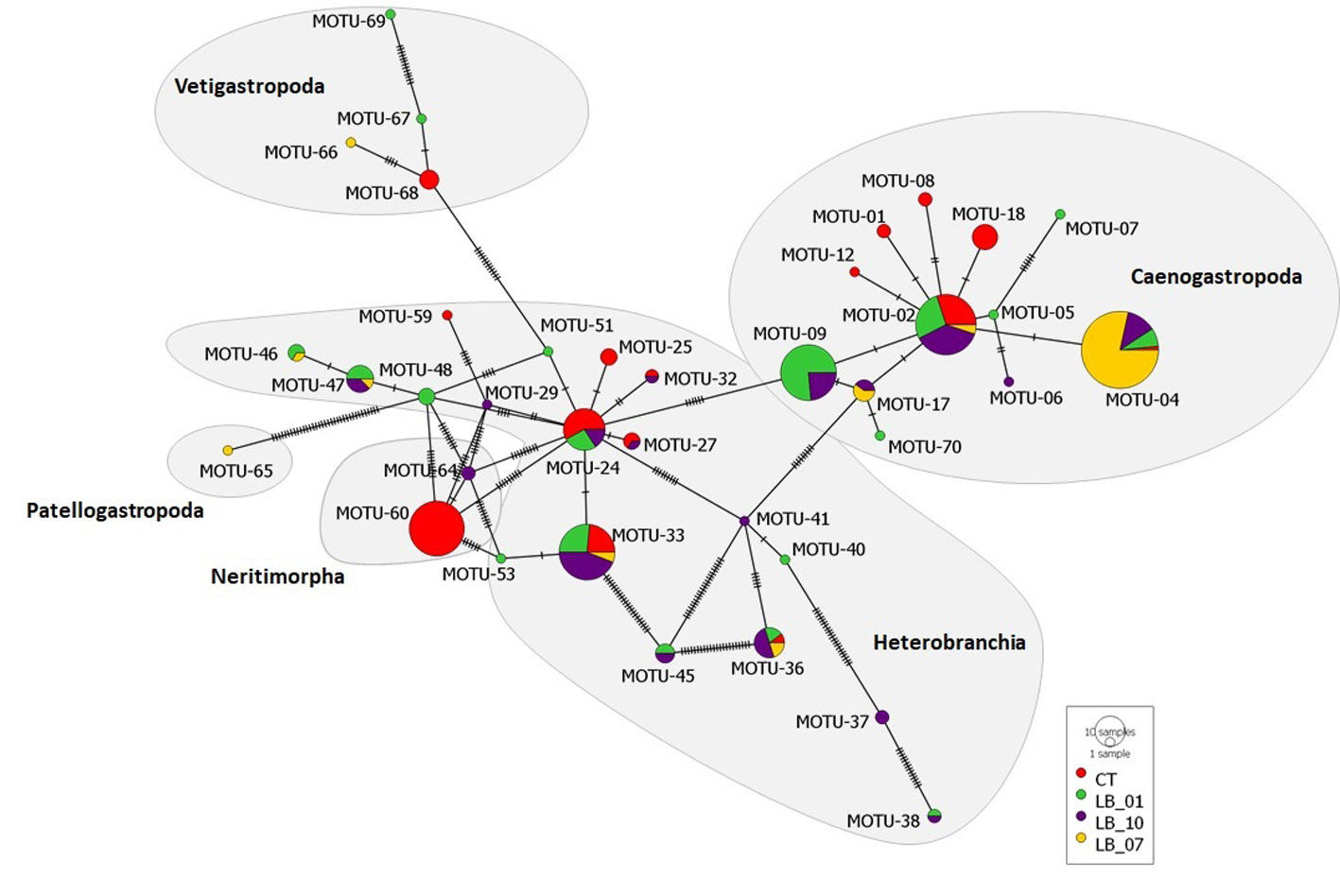
When individual images were organized per MOTU (Supplementary material: Fig. S1[https://doi.org/10.5281/zenodo.7685921]), it was observed that there are many similarities among individuals at distinct taxonomic levels, including subclasses. In contrast, a high genetic diversity was found (nucleotide diversity π = 0.045, with 72 segregating sites and 55 parsimony-informative sites; Supplementary material: Table S1. The phylogenetic analysis inferred from the Minimum Spanning Network indicated that most identified subclasses were represented in all locations (Fig. 3). One exception was Neritimorpha, which was mainly observed in CT.
The MOTUs’ diversity (Shannon index) varied among towing stations with the highest at LB-10 and the lowest at (LB-07) (Fig. 4). This large difference can be inferred by the presence of many MOTU 04 (family Cypraeidae) individuals at LB-07 (Table 2). Even though the diversity was rather similar among CT-01, LB-01, and LB-10 (Fig. 4), the MOTU composition was different with several MOTUs found only in a single towing station (CT-01: 17; LB-01: 16; LB-10: 14; LB-07: 7), i.e., 54 of the 71 MOTUs found were unique (Table 2).

Discussion
A relatively high planktonic gastropod diversity was observed in the present study as compared to that found in 2 sites of the Caribbean Sea: 31 species (Oliva-Rivera & Navarrete, 2007) and 34 species (Chávez-Villegas et al., 2014), but lower than the 62 holoplanktonic species from the Gulf of California (Angulo-Campillo et al., 2011), the 6 species of the order Thecosomata (Sánchez-Hidalgo y Anda, 1989), and the 18 species of the family Atlantidae off the BCP (Aceves-Medina et al., 2020; Moreno-Alcántara et al., 2020). Nevertheless, according to a preliminary rarefaction analysis (results not shown due to reduced sample size) the actual species diversity in our study region is probably under-represented. To overcome the sample size issue, which is mainly due to budget constraints, new research technologies such as environmental DNA (eDNA) metabarcoding are broadening the sampling possibilities (Mychek-Londer et al., 2020).
Differences in community composition between the sampling sites of CT and LB (Fig. 1) would be expected as environmental conditions are usually distinct. Personal observations indicated that both sites are mainly composed of rocky bottoms and a few sandy flats, with a higher abundance of macroalgae at LB than at CT, and clearer waters at CT than LB. These differences, which are probably due to a higher productivity at LB, are explained by the position of the sites relative to the Magdalena Biological Activity Center (MBAC) (Lluch-Belda, 2000), with year-round cooler waters at LB (north of MBAC) than at CT (south of MBAC) (Jerónimo & Gómez-Valdés, 2006). In contrast, the differences in MOTUs composition among stations within LB let us hypothesize that planktonic gastropod distribution within a specific area is probably modulated by micro-environmental processes such as currents and biological events (Hernández-Trujillo et al., 2001). For example, many Cypraeidae individuals at LB-07 points to a massive spawning event in that area.

Even though 18S rDNA has been proposed for biodiversity studies (Ranjithkumar et al., 2018), it generally fails to separate species (Aranceta-Garza et al., 2011; Wu et al., 2015). In addition, the lack of a comprehensive database of gastropod 18S rDNA sequences from the western coast of the Baja California Peninsula limits the capacity of proper species identification. For example, the report of Angulo-Campillo et al. (2011) indicates the presence of several Atlantidae or Desmopteridae species, which in our case could not be distinguished (Table 2). Thus, future studies will require additional genetic markers, such as the 16S rRNA or COI genes (e.g., Crocetta et al., 2020).
The putative presence of molluskan MOTUs not recorded in the study region can be explained by either the low number of 18S rDNA sequences in the public databases, the lack of differences among species in this gene sequence of the same genus or family, and the lack of physical records of those specimens in the study region. In any case, efforts should be devoted to increase the number of gastropod 18S rDNA sequences and other genes in the databases and to promote frequent survey campaigns to detect previously undescribed species in this region. This is particularly relevant because mollusks are gaining attention within climate change research, not only to study phenotypic plasticity and genetic variation (Matoo & Neiman, 2021), but also because of their utility to act as indicators of climatic variability (Molina-González et al., 2018). Also, eDNA metabarcoding, which can be used to detect diverse ecological phenomena such as potential species invasions (e.g., Mychek-Londer et al., 2020), requires solid supporting information.
The presence of non-gastropod sequences within the analyzed individuals is a phenomenon that requires further attention. Even though we cannot rule out the possibility of cross-contamination, these individuals were visualized as planktonic gastropods (Supplementary material: Fig. S1), so it is unlikely that individuals of Cnidaria (order Bivalvulida), and Arthropoda (orders Euphausiacea, and Calanoida) were mistakenly put into the PCR tubes. We also consider that their presence as stomach content or as epibionts was unlikely because when mixed DNA is individually sequenced, it is not possible to separate the signature of each species. Rather, we hypothesize that these sequences might come from eggs or larvae of those other taxa that use gastropod shells as a substratum.
Acknowledgements
Sampling was done under “Pesca de Fomento” permit No. DGOPA.03182.290410.-1591. A. Arciniega received a scholarship from Conacyt (No. 41832). Conacyt provided funds for the project “Conectividad entre poblaciones de moluscos bentónicos marinos, estudio de caso: abulón Haliotis spp.” No. 2007-79482 to R.P.E. Fishermen cooperatives “Meliton Albañez” and “Progreso” supported sampling facilities. J. Cruz Hernández provided technical support during plankton tows. M. C. Rodríguez Jaramillo and R. Inohuye-Rivera provided technical assistance for planktonic gastropod photographic documentation. S. Avila gave technical support with genetic analyses. R. Llera-Herrera provided useful comments.
References
Aceves-Medina, G., Moreno-Alcántara, M., Durazo, R., & Delgado-Hofmann, D. (2020). Distribution of Atlantidae species (Gastropoda: Pterotracheoidea) during an El Niño event in the Southern California Current System (summer-fall 2015). Marine Ecology Progress Series, 648, 153–168. https://doi.org/10.3354/meps13417
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Angulo-Campillo, O., Aceves-Medina, G., & Avedaño-Ibarra, R. (2011). Holoplanktonic mollusks (Mollusca: Gastropoda) from the Gulf of California, México. Check List, 7, 337–342.
Aranceta-Garza, F., Perez-Enriquez, R., & Cruz, P. (2011). PCR-SSCP method for genetic differentiation of canned abalone and commercial gastropods in the Mexican retail market. Food Control, 22, 1015–1020. https://doi.org/10.1016/j.foodcont.2010.11.025
Bandel, K., Riedel, F., & Weikert, H. (1997). Planktonic gastropod larvae from the Red Sea: a synopsis. Ophelia, 47, 151–202.
Bandelt, H., Forster, P., & Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37–48.
Beers, J. R. (1976). Determination of zooplankton biomass. In H. F. Steedman (Ed.), Zooplankton, fixation and preservation. Monographs on oceanographic methodology, No. 4 (pp. 54–60). Paris: The UNESCO Press.
Campos, B., & Landaeta, M. F. (2016). Moluscos planctónicos entre el fiordo Reloncaví y el golfo Corcovado, sur de Chile: ocurrencia, distribución y abundancia en invierno. Revista de Biología Marina y Oceanografía, 51, 527–539. http://dx.doi.org/10.4067/S0718-19572016000300005
Chávez-Villegas, J. F., Enríquez-Díaz, M., & Aldana-Aranda, D. (2014). Abundancia y diversidad larval de gasterópodos en el Caribe Mexicano en relación con la temperatura, la salinidad y el oxígeno disuelto. Revista de Biología Tropical, 62, 223–230.
Courtois de Viçose, G., Viera, M. P., Bilbao, A., & Izquierdo, M. S. (2007). Embryonic and larval development of Haliotis tuberculata coccinea Reeve: an indexed micro-photographic sequence. Journal of Shellfish Research, 26, 847–854. https://doi.org/10.2983/0730-8000(2007)26[847:EALDOH]2.0.CO;2
Crocetta, F., Caputi, L., Paz-Sedano, S., Tanduo, V., Vazzana, A., & Oliverio, M. (2020). High genetic connectivity in a gastropod with long-lived planktonic larvae. Journal of Molluscan Studies, 86, 42–55. https://doi.org/10.1093/mollus/eyz032
Hernández-Trujillo, S., Gómez-Ochoa, F., & Verdugo-Díaz, G. (2001). Dinámica del plancton en la región sur de la Corriente de California. Revista de Biología Tropical, 49, 15–30.
Jerónimo, G., & Gómez-Valdés, J. (2006). Mean temperature and salinity along an isopycnal surface in the upper ocean off Baja California. Ciencias Marinas, 32, 663–671. https://doi.org/10.7773/cm.v32i4.1164
Leigh, J. W. & Bryant, D. (2015). POPART: full-feature software for haplotype network construction. Methods in Ecology and Evolution, 6, 1110–1116. https://doi.org/10.1111/2041-210X.12410
León-Carballo, G., & Muciño-Díaz, M. (1996). Pesquería de abulón. In M. Casas-Valdez, & G. Ponce-Díaz (Eds.), Estudio potencial pesquero y acuícola de Baja California Sur. Vol. I. (pp. 15–41). La Paz, Baja California Sur: Centro de Investigaciones Biológicas del Noroeste, S.C.
Lluch-Belda, D. (2000). Centros de actividad biológica en la costa occidental de Baja California. In D. Lluch-Belda, J. Elorduy-Garay, S. E. Lluch-Cota, & G. Ponce-Díaz (Eds.), BAC: centros de actividad biológica del Pacífico mexicano (pp. 49–64). La Paz, Baja California Sur: Centro de Investigaciones Biológicas del Noroeste, S.C.
Matoo, O. B., & Neiman, M. (2021). Bringing disciplines and people together to characterize the plastic and genetic responses of molluscs to environmental change. Integrative and Comparative Biology, 61, 1689–1698, https://doi.org/10.1093/icb/icab186
Molina-González, O., Lavaniegos, B. E., Gómez-Valdés, J., & De la Cruz-Orozco, M. (2018). Holoplanktonic mollusks off Western Baja California during the weak El Niño 2006-07 and further transition to La Niña. American Malacological Bulletin, 36, 79–95. https://doi.org/10.4003/006.036.0112
Moreno-Alcántara, M., Delgado-Hofmann, D., & Aceves-Medina, G. (2020). Diversity of Atlantidae mollusks (Gastropoda: Pterotracheoidea) from the southern region of the California current off Baja California peninsula, Mexico. Marine Biodiversity, 50, 27. https://doi.org/10.1007/s12526-020-01059-7
Mychek-Londer, J. G., Balasingham, K. D., & Heath, D. D. (2020). Using environmental DNA metabarcoding to map invasive and native invertebrates in two Great Lakes tributaries. Environmental DNA, 2, 283– 297. https://doi.org/10.1002/edn3.56
Oksanen, J., Guillaume-Blanchet, F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D. et al. (2020). Vegan: community ecology package. R package version 2.5-7. Consulted on October 13, 2022 from: https://github.com/vegandevs/vegan
Oliva-Rivera, J. J., & Navarrete, A. J. (2007). Larvas de moluscos gasterópodos del sur de Quintana Roo, México. Hidrobiológica, 17, 151–158
R Core Team (2013). R: a language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria. Consulted on October 13, 2022 from: http://www.R-project.org/
Ranjithkumar, K., Sudhan, C., Utsa, R., & Madhusudhana, R. B. (2018). Ribosomal RNA and their applications in species identification. Journal of Aquaculture in the Tropics, 33, 91–99. https://doi.org/10.32381/JAT.2018.33.01.8
Roegner, G. C. (2000). Transport of molluscan larvae through a shallow estuary. Journal of Plankton Research, 22, 1779–1800. https://doi.org/10.1093/plankt/22.9.1779
Rozas, J., Ferrer-Mata, A., Sánchez-Del Barrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E. et al. (2017). DnaSP v6: DNA sequence polymorphism analysis of large datasets. Molecular Biology and Evolution, 34, 3299–3302. https://doi.org/10.1093/molbev/msx248
Sánchez-Hidalgo y Anda, M. (1989). Gasterópodos holoplanctónicos de la costa occidental de Baja California Sur, en mayo y junio de 1984. Investigaciones Marinas CICIMAR, 4, 1–14.
Schoch, C. (2011). NCBI Taxonomy. Apr 7 [Updated 2020 Feb 11]. In Taxonomy help [Internet]. Bethesda (MD): National Center for Biotechnology Information (US). Consulted on October 13, 2022 from: https://www.ncbi.nlm.nih.gov/books/NBK53758/
Selvamani, M. J. P., Degnan, S. M., & Degnan, B. M. (2001). Microsatellite genotyping of individual abalone larvae: parentage assignment in aquaculture. Marine Biotechnology, 3, 478–485. https://doi.org/10.1007/s1012601-0062-x
Simons, R. A. (2022). ERDDAP. Monterey, CA: NOAA/NMFS/SWFSC/ERD. Consulted on October 13, 2022 from: https://coastwatch.pfeg.noaa.gov/erddap
Smith, P. E., & Richardson, S. L. (1979). Técnicas modelo para prospecciones de huevos de larvas de peces pelágicos. Documento Técnico de Pesca (FAO) spa no. 175. Rome, Italy: FAO.
Wu, S., Xiong, J., & Yu, Y. (2015) Taxonomic resolutions based on 18S rRNA genes: a case study of Subclass Copepoda. Plos One, 10, e0131498. https://doi.org/10.1371/journal.pone.0131498